A new model for SOS-induced mutagenesis: how RecA protein activates DNA polymerase V
- PMID: 20441441
- PMCID: PMC2874081
- DOI: 10.3109/10409238.2010.480968
A new model for SOS-induced mutagenesis: how RecA protein activates DNA polymerase V
Abstract
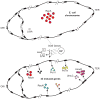
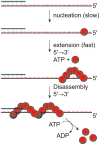
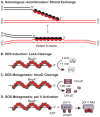
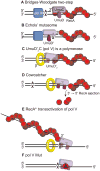
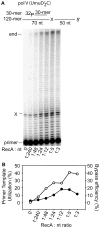
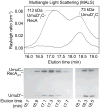
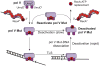
In Escherichia coli, cell survival and genomic stability after UV radiation depends on repair mechanisms induced as part of the SOS response to DNA damage. The early phase of the SOS response is mostly dominated by accurate DNA repair, while the later phase is characterized with elevated mutation levels caused by error-prone DNA replication. SOS mutagenesis is largely the result of the action of DNA polymerase V (pol V), which has the ability to insert nucleotides opposite various DNA lesions in a process termed translesion DNA synthesis (TLS). Pol V is a low-fidelity polymerase that is composed of UmuD'(2)C and is encoded by the umuDC operon. Pol V is strictly regulated in the cell so as to avoid genomic mutation overload. RecA nucleoprotein filaments (RecA*), formed by RecA binding to single-stranded DNA with ATP, are essential for pol V-catalyzed TLS both in vivo and in vitro. This review focuses on recent studies addressing the protein composition of active DNA polymerase V, and the role of RecA protein in activating this enzyme. Based on unforeseen properties of RecA*, we describe a new model for pol V-catalyzed SOS-induced mutagenesis.
Figures



Similar articles
-
The active form of DNA polymerase V is UmuD'(2)C-RecA-ATP.Nature. 2009 Jul 16;460(7253):359-63. doi: 10.1038/nature08178. Nature. 2009. PMID: 19606142 Free PMC article.
-
DNA polymerase V and RecA protein, a minimal mutasome.Mol Cell. 2005 Feb 18;17(4):561-72. doi: 10.1016/j.molcel.2005.01.006. Mol Cell. 2005. PMID: 15721259
-
Visualization of two binding sites for the Escherichia coli UmuD'(2)C complex (DNA pol V) on RecA-ssDNA filaments.J Mol Biol. 2000 Mar 31;297(3):585-97. doi: 10.1006/jmbi.2000.3591. J Mol Biol. 2000. PMID: 10731413
-
Mutations for Worse or Better: Low-Fidelity DNA Synthesis by SOS DNA Polymerase V Is a Tightly Regulated Double-Edged Sword.Biochemistry. 2016 Apr 26;55(16):2309-18. doi: 10.1021/acs.biochem.6b00117. Epub 2016 Apr 12. Biochemistry. 2016. PMID: 27043933 Free PMC article. Review.
-
A Comprehensive View of Translesion Synthesis in Escherichia coli.Microbiol Mol Biol Rev. 2020 Jun 17;84(3):e00002-20. doi: 10.1128/MMBR.00002-20. Print 2020 Aug 19. Microbiol Mol Biol Rev. 2020. PMID: 32554755 Free PMC article. Review.
Cited by
-
The SOS system: A complex and tightly regulated response to DNA damage.Environ Mol Mutagen. 2019 May;60(4):368-384. doi: 10.1002/em.22267. Epub 2019 Jan 7. Environ Mol Mutagen. 2019. PMID: 30447030 Free PMC article. Review.
-
Recombinational branch migration by the RadA/Sms paralog of RecA in Escherichia coli.Elife. 2016 Feb 4;5:e10807. doi: 10.7554/eLife.10807. Elife. 2016. PMID: 26845522 Free PMC article.
-
Suppression of the E. coli SOS response by dNTP pool changes.Nucleic Acids Res. 2015 Apr 30;43(8):4109-20. doi: 10.1093/nar/gkv217. Epub 2015 Mar 30. Nucleic Acids Res. 2015. PMID: 25824947 Free PMC article.
-
New insights into the structures and interactions of bacterial Y-family DNA polymerases.Nucleic Acids Res. 2019 May 21;47(9):4393-4405. doi: 10.1093/nar/gkz198. Nucleic Acids Res. 2019. PMID: 30916324 Free PMC article.
-
Creating directed double-strand breaks with the Ref protein: a novel RecA-dependent nuclease from bacteriophage P1.J Biol Chem. 2011 Mar 11;286(10):8240-8251. doi: 10.1074/jbc.M110.205088. Epub 2010 Dec 30. J Biol Chem. 2011. PMID: 21193392 Free PMC article.
References
-
- Arenson TA, Tsodikov OV, Cox MM. Quantitative analysis of the kinetics of end-dependent disassembly of RecA filaments from ssDNA. J Mol Biol. 1999;288:391–401. - PubMed
-
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–4. - PubMed
-
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–56. - PubMed
-
- Bork JM, Cox MM, Inman RB. RecA protein filaments disassemble in the 5′ to 3′ direction on single-stranded DNA. Journal of Biological Chemistry. 2001;276:45740–43. - PubMed
-
- Boudsocq F, Campbell M, Devoret R, Bailone A. Quantitation of the inhibition of Hfr × F- recombination by the mutagenesis complex UmuD′C. J Mol Biol. 1997;270:201–11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
