Direct sequencing of the human microbiome readily reveals community differences
- PMID: 20441597
- PMCID: PMC2898070
- DOI: 10.1186/gb-2010-11-5-210
Direct sequencing of the human microbiome readily reveals community differences
Abstract
Culture-independent studies of human microbiota by direct genomic sequencing reveal quite distinct differences among communities, indicating that improved sequencing capacity can be most wisely utilized to study more samples, rather than more sequences per sample.
Figures

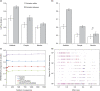

References
-
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J. et al.Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. - PMC - PubMed

