Linking well-tempered metadynamics simulations with experiments
- PMID: 20441734
- PMCID: PMC2862196
- DOI: 10.1016/j.bpj.2010.01.033
Linking well-tempered metadynamics simulations with experiments
Abstract
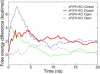
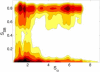
Linking experiments with the atomistic resolution provided by molecular dynamics simulations can shed light on the structure and dynamics of protein-disordered states. The sampling limitations of classical molecular dynamics can be overcome using metadynamics, which is based on the introduction of a history-dependent bias on a small number of suitably chosen collective variables. Even if such bias distorts the probability distribution of the other degrees of freedom, the equilibrium Boltzmann distribution can be reconstructed using a recently developed reweighting algorithm. Quantitative comparison with experimental data is thus possible. Here we show the potential of this combined approach by characterizing the conformational ensemble explored by a 13-residue helix-forming peptide by means of a well-tempered metadynamics/parallel tempering approach and comparing the reconstructed nuclear magnetic resonance scalar couplings with experimental data.
Copyright (c) 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Reconstructing the equilibrium Boltzmann distribution from well-tempered metadynamics.J Comput Chem. 2009 Aug;30(11):1615-21. doi: 10.1002/jcc.21305. J Comput Chem. 2009. PMID: 19421997
-
Probing the Conformational Ensemble of the Amyloid Beta 16-22 Fragment with Parallel-Bias Metadynamics.J Phys Chem B. 2024 Dec 19;128(50):12333-12347. doi: 10.1021/acs.jpcb.4c04919. Epub 2024 Dec 5. J Phys Chem B. 2024. PMID: 39635892
-
Essential slow degrees of freedom in protein-surface simulations: A metadynamics investigation.Biochem Biophys Res Commun. 2018 Mar 29;498(2):274-281. doi: 10.1016/j.bbrc.2017.07.066. Epub 2017 Jul 15. Biochem Biophys Res Commun. 2018. PMID: 28720500
-
Molecular Dynamics Simulations Combined with Nuclear Magnetic Resonance and/or Small-Angle X-ray Scattering Data for Characterizing Intrinsically Disordered Protein Conformational Ensembles.J Chem Inf Model. 2019 May 28;59(5):1743-1758. doi: 10.1021/acs.jcim.8b00928. Epub 2019 Mar 18. J Chem Inf Model. 2019. PMID: 30840442 Review.
-
Computational studies of transport in ion channels using metadynamics.Biochim Biophys Acta. 2016 Jul;1858(7 Pt B):1733-40. doi: 10.1016/j.bbamem.2016.02.015. Epub 2016 Feb 15. Biochim Biophys Acta. 2016. PMID: 26891818 Review.
Cited by
-
Molecular dynamics studies reveal structural and functional features of the SARS-CoV-2 spike protein.Bioessays. 2022 Sep;44(9):e2200060. doi: 10.1002/bies.202200060. Epub 2022 Jul 17. Bioessays. 2022. PMID: 35843871 Free PMC article. Review.
-
The influence of antibody humanization on shark variable domain (VNAR) binding site ensembles.Front Immunol. 2022 Sep 2;13:953917. doi: 10.3389/fimmu.2022.953917. eCollection 2022. Front Immunol. 2022. PMID: 36177031 Free PMC article.
-
Molecular View on the iRGD Peptide Binding Mechanism: Implications for Integrin Activity and Selectivity Profiles.J Chem Inf Model. 2023 Oct 23;63(20):6302-6315. doi: 10.1021/acs.jcim.3c01071. Epub 2023 Oct 3. J Chem Inf Model. 2023. PMID: 37788340 Free PMC article.
-
Structural Characterization of Nanobodies during Germline Maturation.Biomolecules. 2023 Feb 17;13(2):380. doi: 10.3390/biom13020380. Biomolecules. 2023. PMID: 36830754 Free PMC article.
-
Exploring the mechanism of zanamivir resistance in a neuraminidase mutant: a molecular dynamics study.PLoS One. 2012;7(9):e44057. doi: 10.1371/journal.pone.0044057. Epub 2012 Sep 6. PLoS One. 2012. PMID: 22970161 Free PMC article.
References
-
- Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. - PubMed
-
- Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. - PubMed
-
- Vendruscolo M. Determination of conformationally heterogeneous states of proteins. Curr. Opin. Struct. Biol. 2007;17:15–20. - PubMed
-
- Barducci A., Bussi G., Parrinello M. Well-tempered metadynamics: a smoothly converging and tunable free-energy method. Phys. Rev. Lett. 2008;100:020603. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

