Prediction of protein-protein interaction sites using electrostatic desolvation profiles
- PMID: 20441756
- PMCID: PMC2862153
- DOI: 10.1016/j.bpj.2009.12.4332
Prediction of protein-protein interaction sites using electrostatic desolvation profiles
Abstract
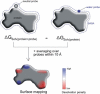
Protein-protein complex formation involves removal of water from the interface region. Surface regions with a small free energy penalty for water removal or desolvation may correspond to preferred interaction sites. A method to calculate the electrostatic free energy of placing a neutral low-dielectric probe at various protein surface positions has been designed and applied to characterize putative interaction sites. Based on solutions of the finite-difference Poisson equation, this method also includes long-range electrostatic contributions and the protein solvent boundary shape in contrast to accessible-surface-area-based solvation energies. Calculations on a large set of proteins indicate that in many cases (>90%), the known binding site overlaps with one of the six regions of lowest electrostatic desolvation penalty (overlap with the lowest desolvation region for 48% of proteins). Since the onset of electrostatic desolvation occurs even before direct protein-protein contact formation, it may help guide proteins toward the binding region in the final stage of complex formation. It is interesting that the probe desolvation properties associated with residue types were found to depend to some degree on whether the residue was outside of or part of a binding site. The probe desolvation penalty was on average smaller if the residue was part of a binding site compared to other surface locations. Applications to several antigen-antibody complexes demonstrated that the approach might be useful not only to predict protein interaction sites in general but to map potential antigenic epitopes on protein surfaces.
Copyright (c) 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Computational antigenic epitope prediction by calculating electrostatic desolvation penalties of protein surfaces.Methods Mol Biol. 2014;1184:365-74. doi: 10.1007/978-1-4939-1115-8_20. Methods Mol Biol. 2014. PMID: 25048135
-
Solvation energy density occlusion approximation for evaluation of desolvation penalties in biomolecular interactions.Proteins. 2001 Apr 1;43(1):12-27. doi: 10.1002/1097-0134(20010401)43:1<12::aid-prot1013>3.0.co;2-7. Proteins. 2001. PMID: 11170210
-
Free energy landscapes of encounter complexes in protein-protein association.Biophys J. 1999 Mar;76(3):1166-78. doi: 10.1016/S0006-3495(99)77281-4. Biophys J. 1999. PMID: 10049302 Free PMC article.
-
Protein electrostatics: a review of the equations and methods used to model electrostatic equations in biomolecules--applications in biotechnology.Biotechnol Annu Rev. 2003;9:315-95. doi: 10.1016/s1387-2656(03)09010-0. Biotechnol Annu Rev. 2003. PMID: 14650935 Review.
-
Prediction of the binding energy for small molecules, peptides and proteins.J Mol Recognit. 1999 May-Jun;12(3):177-90. doi: 10.1002/(SICI)1099-1352(199905/06)12:3<177::AID-JMR451>3.0.CO;2-Z. J Mol Recognit. 1999. PMID: 10398408 Review.
Cited by
-
SPPS: a sequence-based method for predicting probability of protein-protein interaction partners.PLoS One. 2012;7(1):e30938. doi: 10.1371/journal.pone.0030938. Epub 2012 Jan 26. PLoS One. 2012. PMID: 22292078 Free PMC article.
-
Protein Interaction with Charged Macromolecules: From Model Polymers to Unfolded Proteins and Post-Translational Modifications.Int J Mol Sci. 2019 Mar 12;20(5):1252. doi: 10.3390/ijms20051252. Int J Mol Sci. 2019. PMID: 30871103 Free PMC article. Review.
-
Structural and Computational Biology in the Design of Immunogenic Vaccine Antigens.J Immunol Res. 2015;2015:156241. doi: 10.1155/2015/156241. Epub 2015 Oct 7. J Immunol Res. 2015. PMID: 26526043 Free PMC article. Review.
-
Sequence- and Structure-Based Immunoreactive Epitope Discovery for Burkholderia pseudomallei Flagellin.PLoS Negl Trop Dis. 2015 Jul 29;9(7):e0003917. doi: 10.1371/journal.pntd.0003917. eCollection 2015. PLoS Negl Trop Dis. 2015. PMID: 26222657 Free PMC article.
-
Algorithmic approaches to protein-protein interaction site prediction.Algorithms Mol Biol. 2015 Feb 15;10:7. doi: 10.1186/s13015-015-0033-9. eCollection 2015. Algorithms Mol Biol. 2015. PMID: 25713596 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

