Quantifying the labeling and the levels of plant cell wall precursors using ion chromatography tandem mass spectrometry
- PMID: 20442274
- PMCID: PMC2899904
- DOI: 10.1104/pp.110.155713
Quantifying the labeling and the levels of plant cell wall precursors using ion chromatography tandem mass spectrometry
Abstract
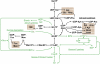
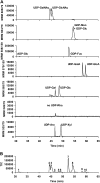
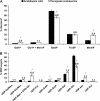
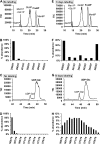
The biosynthesis of cell wall polymers involves enormous fluxes through central metabolism that are not fully delineated and whose regulation is poorly understood. We have established and validated a liquid chromatography tandem mass spectrometry method using multiple reaction monitoring mode to separate and quantify the levels of plant cell wall precursors. Target analytes were identified by their parent/daughter ions and retention times. The method allows the quantification of precursors at low picomole quantities with linear responses up to the nanomole quantity range. When applying the technique to Arabidopsis (Arabidopsis thaliana) T87 cell cultures, 16 hexose-phosphates (hexose-Ps) and nucleotide-sugars (NDP-sugars) involved in cell wall biosynthesis were separately quantified. Using hexose-P and NDP-sugar standards, we have shown that hot water extraction allows good recovery of the target metabolites (over 86%). This method is applicable to quantifying the levels of hexose-Ps and NDP-sugars in different plant tissues, such as Arabidopsis T87 cells in culture and fenugreek (Trigonella foenum-graecum) endosperm tissue, showing higher levels of galacto-mannan precursors in fenugreek endosperm. In Arabidopsis cells incubated with [U-(13)C(Fru)]sucrose, the method was used to track the labeling pattern in cell wall precursors. As the fragmentation of hexose-Ps and NDP-sugars results in high yields of [PO(3)](-)/or [H(2)PO(4)](-) ions, mass isotopomers can be quantified directly from the intensity of selected tandem mass spectrometry transitions. The ability to directly measure (13)C labeling in cell wall precursors makes possible metabolic flux analysis of cell wall biosynthesis based on dynamic labeling experiments.
Figures

References
-
- Allwood JW, Goodacre R. (2010) An introduction to liquid chromatography-mass spectrometry instrumentation applied in plant metabolomic analyses. Phytochem Anal 21: 33–47 - PubMed
-
- Arrivault S, Guenther M, Ivakov A, Feil R, Vosloh D, van Dongen JT, Sulpice R, Stitt M. (2009) Use of reverse-phase liquid chromatography, linked to tandem mass spectrometry, to profile the Calvin cycle and other metabolic intermediates in Arabidopsis rosettes at different carbon dioxide concentrations. Plant J 59: 824–839 - PubMed
-
- Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. (2006) Separation and quantification of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr A 1125: 76–88 - PubMed
-
- Canelas AB, ten Pierick A, Ras C, Seifar RM, van Dam JC, van Gulik WM, Heijnen JJ. (2009) Quantitative evaluation of intracellular metabolite extraction techniques for yeast metabolomics. Anal Chem 81: 7379–7389 - PubMed
-
- Cruz JA, Emery C, Wüst M, Kramer DM, Lange BM. (2008) Metabolite profiling of Calvin cycle intermediates by HPLC-MS using mixed-mode stationary phases. Plant J 55: 1047–1060 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

