LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis
- PMID: 20442277
- PMCID: PMC2899912
- DOI: 10.1104/pp.110.157446
LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis
Abstract
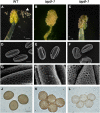
Pollen grains of land plants have evolved remarkably strong outer walls referred to as exine that protect pollen and interact with female stigma cells. Exine is composed of sporopollenin, and while the composition and synthesis of this biopolymer are not well understood, both fatty acids and phenolics are likely components. Here, we describe mutations in the Arabidopsis (Arabidopsis thaliana) LESS ADHESIVE POLLEN (LAP5) and LAP6 that affect exine development. Mutation of either gene results in abnormal exine patterning, whereas pollen of double mutants lacked exine deposition and subsequently collapsed, causing male sterility. LAP5 and LAP6 encode anther-specific proteins with homology to chalcone synthase, a key flavonoid biosynthesis enzyme. lap5 and lap6 mutations reduced the accumulation of flavonoid precursors and flavonoids in developing anthers, suggesting a role in the synthesis of phenolic constituents of sporopollenin. Our in vitro functional analysis of LAP5 and LAP6 using 4-coumaroyl-coenzyme A yielded bis-noryangonin (a commonly reported derailment product of chalcone synthase), while similar in vitro analyses using fatty acyl-coenzyme A as the substrate yielded medium-chain alkyl pyrones. Thus, in vitro assays indicate that LAP5 and LAP6 are multifunctional enzymes and may play a role in both the synthesis of pollen fatty acids and phenolics found in exine. Finally, the genetic interaction between LAP5 and an anther gene involved in fatty acid hydroxylation (CYP703A2) demonstrated that they act synergistically in exine production.
Figures









Similar articles
-
CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis.Plant Physiol. 2009 Oct;151(2):574-89. doi: 10.1104/pp.109.144469. Epub 2009 Aug 21. Plant Physiol. 2009. PMID: 19700560 Free PMC article.
-
LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl α-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana.Plant Cell. 2010 Dec;22(12):4045-66. doi: 10.1105/tpc.110.080028. Epub 2010 Dec 30. Plant Cell. 2010. PMID: 21193570 Free PMC article.
-
ABCG26-mediated polyketide trafficking and hydroxycinnamoyl spermidines contribute to pollen wall exine formation in Arabidopsis.Plant Cell. 2014 Nov;26(11):4483-98. doi: 10.1105/tpc.114.130484. Epub 2014 Nov 21. Plant Cell. 2014. PMID: 25415974 Free PMC article.
-
The biosynthesis, composition and assembly of the outer pollen wall: A tough case to crack.Phytochemistry. 2015 May;113:170-82. doi: 10.1016/j.phytochem.2014.05.002. Epub 2014 Jun 3. Phytochemistry. 2015. PMID: 24906292 Review.
-
Genetic regulation of sporopollenin synthesis and pollen exine development.Annu Rev Plant Biol. 2011;62:437-60. doi: 10.1146/annurev-arplant-042809-112312. Annu Rev Plant Biol. 2011. PMID: 21275644 Review.
Cited by
-
Different expression pattern of flowering pathway genes contribute to male or female organ development during floral transition in the monoecious weed Ambrosia artemisiifolia L. (Asteraceae).PeerJ. 2019 Oct 4;7:e7421. doi: 10.7717/peerj.7421. eCollection 2019. PeerJ. 2019. PMID: 31598422 Free PMC article.
-
Distinctive Gene Expression Patterns Define Endodormancy to Ecodormancy Transition in Apricot and Peach.Front Plant Sci. 2020 Feb 28;11:180. doi: 10.3389/fpls.2020.00180. eCollection 2020. Front Plant Sci. 2020. PMID: 32180783 Free PMC article.
-
Fertile Arabidopsis cyp704b1 mutant, defective in sporopollenin biosynthesis, has a normal pollen coat and lipidic organelles in the tapetum.Plant Biotechnol (Tokyo). 2021 Mar 25;38(1):109-116. doi: 10.5511/plantbiotechnology.20.1214b. Plant Biotechnol (Tokyo). 2021. PMID: 34177330 Free PMC article.
-
PpASCL, the Physcomitrella patens Anther-Specific Chalcone Synthase-Like Enzyme Implicated in Sporopollenin Biosynthesis, Is Needed for Integrity of the Moss Spore Wall and Spore Viability.PLoS One. 2016 Jan 11;11(1):e0146817. doi: 10.1371/journal.pone.0146817. eCollection 2016. PLoS One. 2016. PMID: 26752629 Free PMC article.
-
Identification of Sporopollenin as the Outer Layer of Cell Wall in Microalga Chlorella protothecoides.Front Microbiol. 2016 Jun 30;7:1047. doi: 10.3389/fmicb.2016.01047. eCollection 2016. Front Microbiol. 2016. PMID: 27446068 Free PMC article.
References
-
- Aarts MG, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, Stiekema WJ, Scott R, Pereira A. (1997) The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J 12: 615–623 - PubMed
-
- Ageez A, Kazama Y, Sugiyama R, Kawano S. (2005) Male-fertility genes expressed in male flower buds of Silene latifolia include homologs of anther-specific genes. Genes Genet Syst 80: 403–413 - PubMed
-
- Ahlers F, Thom I, Lambert J, Kuckuk R, Wiermann R. (1999) 1H NMR analysis of sporopollenin from Typha angustifolia. Phytochemistry 5: 1095–1098
-
- Akiyama T, Shibuya M, Liu HM, Ebizuka Y. (1999) p-Coumaroyltriacetic acid synthase, a new homologue of chalcone synthase, from Hydrangea macrophylla var. thunbergii. Eur J Biochem 263: 834–839 - PubMed
-
- Ariizumi T, Hatakeyama K, Hinata K, Sato S, Kato T, Tabata S, Toriyama K. (2003) A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol Biol 53: 107–116 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

