mRNA and microRNA expression profiles of the NCI-60 integrated with drug activities
- PMID: 20442302
- PMCID: PMC2879615
- DOI: 10.1158/1535-7163.MCT-09-0965
mRNA and microRNA expression profiles of the NCI-60 integrated with drug activities
Abstract
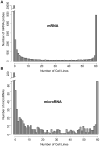
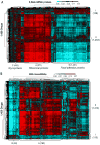
As part of the Spotlight on Molecular Profiling series, we present here new profiling studies of mRNA and microRNA expression for the 60 cell lines of the National Cancer Institute (NCI) Developmental Therapeutics program (DTP) drug screen (NCI-60) using the 41,000-probe Agilent Whole Human Genome Oligo Microarray and the 15,000-feature Agilent Human microRNA Microarray V2. The expression levels of approximately 21,000 genes and 723 human microRNAs were measured. These profiling studies include quadruplicate technical replicates for six and eight cell lines for mRNA and microRNA, respectively, and duplicates for the remaining cell lines. The resulting data sets are freely available and searchable online in our CellMiner database. The result indicates high reproducibility for both platforms and an essential biological similarity across the various cell types. The mRNA and microRNA expression levels were integrated with our previously published 1,429-compound database of anticancer activity obtained from the NCI DTP drug screen. Large blocks of both mRNAs and microRNAs were identified with predominately unidirectional correlations to approximately 1,300 drugs, including 121 drugs with known mechanisms of action. The data sets presented here will facilitate the identification of groups of mRNAs, microRNAs, and drugs that potentially affect and interact with one another.
Conflict of interest statement
No conflicts of interest to disclose.
Figures



References
-
- Jay C, Nemunaitis J, Chen P, Fulgham P, Tong AW. mi RNA Profiling for Diagnosis and Prognosis of Human Cancer. DNA and Cell Biology. 2007;26:293–300. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. - PubMed
-
- Aravin A, Tuschl T. Identification and characterization of small RNAs involved in RNA silencing. FEBS Lett. 2005;579:5830–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

