Alteration of substrate specificity: the variable N-terminal domain of tobacco Ca(2+)-dependent protein kinase is important for substrate recognition
- PMID: 20442373
- PMCID: PMC2899867
- DOI: 10.1105/tpc.109.073577
Alteration of substrate specificity: the variable N-terminal domain of tobacco Ca(2+)-dependent protein kinase is important for substrate recognition
Abstract
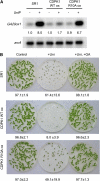
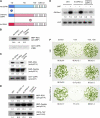
Protein kinases are major signaling molecules that are involved in a variety of cellular processes. However, the molecular mechanisms whereby protein kinases discriminate specific substrates are still largely unknown. Ca(2+)-dependent protein kinases (CDPKs) play central roles in Ca(2+) signaling in plants. Previously, we found that a tobacco (Nicotiana tabacum) CDPK1 negatively regulated the transcription factor REPRESSION OF SHOOT GROWTH (RSG), which is involved in gibberellin feedback regulation. Here, we found that the variable N-terminal domain of CDPK1 is necessary for the recognition of RSG. A mutation (R10A) in the variable N-terminal domain of CDPK1 reduced both RSG binding and RSG phosphorylation while leaving kinase activity intact. Furthermore, the R10A mutation suppressed the in vivo function of CDPK1. The substitution of the variable N-terminal domain of an Arabidopsis thaliana CDPK, At CPK9, with that of Nt CDPK1 conferred RSG kinase activities. This chimeric CDPK behaved according to the identity of the variable N-terminal domain in transgenic plants. Our results open the possibility of engineering the substrate specificity of CDPK by manipulation of the variable N-terminal domain, enabling a rational rewiring of cellular signaling pathways.
Figures



References
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
-
- Asano T., Tanaka N., Yang G., Hayashi N., Komatsu S. (2005). Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: Comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol. 46: 356–366 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

