Isotopologue profiling of Legionella pneumophila: role of serine and glucose as carbon substrates
- PMID: 20442401
- PMCID: PMC2903384
- DOI: 10.1074/jbc.M110.128678
Isotopologue profiling of Legionella pneumophila: role of serine and glucose as carbon substrates
Abstract
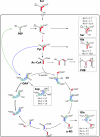
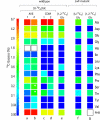
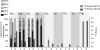
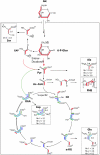
Legionella pneumophila (Lp) is commonly found in freshwater habitats but is also the causative agent of Legionnaires' disease when infecting humans. Although various virulence factors have been reported, little is known about the nutrition and the metabolism of the bacterium. Here, we report the application of isotopologue profiling for analyzing the metabolism of L. pneumophila. Cultures of Lp were supplied with [U-(13)C(3)]serine, [U-(13)C(6)]glucose, or [1,2-(13)C(2)]glucose. After growth, (13)C enrichments and isotopologue patterns of protein-derived amino acids and poly-3-hydroxybutyrate were determined by mass spectrometry and/or NMR spectroscopy. The labeling patterns detected in the experiment with [U-(13)C(3)]serine showed major carbon flux from serine to pyruvate and from pyruvate to acetyl-CoA, which serves as a precursor of poly-3-hydroxybutyrate or as a substrate of a complete citrate cycle with Si specificity of the citrate synthase. Minor carbon flux was observed between pyruvate and oxaloacetate/malate by carboxylation and decarboxylation, respectively. The apparent lack of label in Val, Ile, Leu, Pro, Phe, Met, Arg, and Tyr confirmed that L. pneumophila is auxotrophic for these amino acids. Experiments with [(13)C]glucose showed that the carbohydrate is also used as a substrate to feed the central metabolism. The specific labeling patterns due to [1,2-(13)C(2)]glucose identified the Entner-Doudoroff pathway as the predominant route for glucose utilization. In line with these observations, a mutant lacking glucose-6-phosphate dehydrogenase (Delta zwf) did not incorporate label from glucose at significant levels and was slowly outcompeted by the wild type strain in successive rounds of infection in Acanthamoeba castellanii, indicating the importance of this enzyme and of carbohydrate usage in general for the life cycle of Lp.
Figures
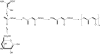
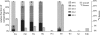


References
-
- Hammer B. K., Tateda E. S., Swanson M. S. (2002) Mol. Microbiol. 44, 107–118 - PubMed
-
- Molofsky A. B., Swanson M. S. (2003) Mol. Microbiol. 50, 445–461 - PubMed
-
- Molofsky A. B., Swanson M. S. (2004) Mol. Microbiol. 53, 29–40 - PubMed
-
- Swanson M. S., Hammer B. K. (2000) Annu. Rev. Microbiol. 54, 567–613 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

