Systems approaches to polypharmacology and drug discovery
- PMID: 20443163
- PMCID: PMC3068535
Systems approaches to polypharmacology and drug discovery
Abstract
Systems biology uses experimental and computational approaches to characterize large sample populations systematically, process large datasets, examine and analyze regulatory networks, and model reactions to determine how components are joined to form functional systems. Systems biology technologies, data and knowledge are particularly useful in understanding disease processes and drug actions. An important area of integration between systems biology and drug discovery is the concept of polypharmacology: the treatment of diseases by modulating more than one target. Polypharmacology for complex diseases is likely to involve multiple drugs acting on distinct targets that are part of a network regulating physiological responses. This review discusses the current state of the systems-level understanding of diseases and both the therapeutic and adverse mechanisms of drug actions. Drug-target networks can be used to identify multiple targets and to determine suitable combinations of drug targets or drugs. Thus, the discovery of new drug therapies for complex diseases may be greatly aided by systems biology.
Figures

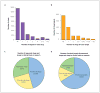
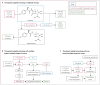


References
-
- Bamborough P, Drewry D, Harper G, Smith GK, Schneider K. Assessment of chemical coverage of kinome space and its implications for kinase drug discovery. J Med Chem. 2008;51 (24):7898–7914. - PubMed
-
- Frye SV. Structure-activity relationship homology (SARAH): A conceptual framework for drug discovery in the genomic era. Chem Biol. 1999;6(1):R3–R7. - PubMed
-
- Lovering F, Bikker J, Humblet C. Escape from flatland: Increasing saturation as an approach to improving clinical success. J Med Chem. 2009;52(21):6752–6756. - PubMed
-
- Gordian M, Singh N, Zemmel R, Elias T. Why products fail in phase III. In Vivo. 2006:2006800066.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
