Analysis of genetic diversity in banana cultivars (Musa cvs.) from the South of Oman using AFLP markers and classification by phylogenetic, hierarchical clustering and principal component analyses
- PMID: 20443211
- PMCID: PMC2865835
- DOI: 10.1631/jzus.B0900310
Analysis of genetic diversity in banana cultivars (Musa cvs.) from the South of Oman using AFLP markers and classification by phylogenetic, hierarchical clustering and principal component analyses
Abstract
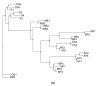
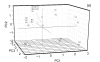
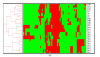
Banana is an important crop grown in Oman and there is a dearth of information on its genetic diversity to assist in crop breeding and improvement programs. This study employed amplified fragment length polymorphism (AFLP) to investigate the genetic variation in local banana cultivars from the southern region of Oman. Using 12 primer combinations, a total of 1094 bands were scored, of which 1012 were polymorphic. Eighty-two unique markers were identified, which revealed the distinct separation of the seven cultivars. The results obtained show that AFLP can be used to differentiate the banana cultivars. Further classification by phylogenetic, hierarchical clustering and principal component analyses showed significant differences between the clusters found with molecular markers and those clusters created by previous studies using morphological analysis. Based on the analytical results, a consensus dendrogram of the banana cultivars is presented.
Figures






Similar articles
-
Musa genetic diversity revealed by SRAP and AFLP.Mol Biotechnol. 2011 Mar;47(3):189-99. doi: 10.1007/s12033-010-9328-8. Mol Biotechnol. 2011. PMID: 20803102
-
Amplified fragment length polymorphism fingerprinting of 16 banana cultivars (Musa cvs.).Mol Phylogenet Evol. 2000 Dec;17(3):360-6. doi: 10.1006/mpev.2000.0848. Mol Phylogenet Evol. 2000. PMID: 11133190
-
Assessment of the validity of the sections in Musa (musaceae) using AFLP.Ann Bot. 2002 Aug;90(2):231-8. doi: 10.1093/aob/mcf170. Ann Bot. 2002. PMID: 12197520 Free PMC article.
-
Determination of genetic relationships between evergreen azalea cultivars in China using AFLP markers.J Zhejiang Univ Sci B. 2013 Apr;14(4):299-308. doi: 10.1631/jzus.B1200094. J Zhejiang Univ Sci B. 2013. PMID: 23549847 Free PMC article.
-
Advances in Somatic Embryogenesis of Banana.Int J Mol Sci. 2023 Jul 1;24(13):10999. doi: 10.3390/ijms241310999. Int J Mol Sci. 2023. PMID: 37446177 Free PMC article. Review.
Cited by
-
Narrow genetic diversity in germplasm from the Guinean and Sudano-Guinean zones in Benin indicates the need to broaden the genetic base of sweet fig banana (Musa acuminata cv Sotoumon).PLoS One. 2023 Nov 16;18(11):e0294315. doi: 10.1371/journal.pone.0294315. eCollection 2023. PLoS One. 2023. PMID: 37972084 Free PMC article.
-
Molecular identification and evolutionary relationships between the subspecies of Musa by DNA barcodes.BMC Genomics. 2020 Sep 24;21(1):659. doi: 10.1186/s12864-020-07036-5. BMC Genomics. 2020. PMID: 32972362 Free PMC article.
-
Seed Oil Quality and Cultivation of Sambucus williamsii Hance as a New Oil Crop.Front Nutr. 2021 Dec 24;8:796175. doi: 10.3389/fnut.2021.796175. eCollection 2021. Front Nutr. 2021. PMID: 35004823 Free PMC article.
-
The Promise of Molecular and Genomic Techniques for Biodiversity Research and DNA Barcoding of the Arabian Peninsula Flora.Front Plant Sci. 2019 Jan 21;9:1929. doi: 10.3389/fpls.2018.01929. eCollection 2018. Front Plant Sci. 2019. PMID: 30719028 Free PMC article. Review.
-
The triploid East African Highland Banana (EAHB) genepool is genetically uniform arising from a single ancestral clone that underwent population expansion by vegetative propagation.Theor Appl Genet. 2016 Mar;129(3):547-61. doi: 10.1007/s00122-015-2647-1. Epub 2016 Jan 8. Theor Appl Genet. 2016. PMID: 26743524
References
-
- Bhat KV, Amaravathi Y, Guatam PL, Velayudhan KC. AFLP characterization and genetic diversity analysis of Indian banana and plantain cultivars (Musa spp.) Plant Genetic Resources. 2004;2(2):121–130. doi: 10.1079/PGR200440. - DOI
-
- de Langhe E. Banana Diversity in the Middle East (Jordan, Egypt and Oman) Montpellier, France: International Network for the Improvement of Banana and Plantain (INIBAP); 2002.
-
- Doyle JJ, Doyle JL. A rapid total DNA preparation procedure for fresh plant tissue. Focus. 1990;12:13–15.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

