Characterization of the structures of phosphodiesterase 10 binding with adenosine 3',5'-monophosphate and guanosine 3',5'-monophosphate by hybrid quantum mechanical/molecular mechanical calculations
- PMID: 20443609
- PMCID: PMC2878663
- DOI: 10.1021/jp911527y
Characterization of the structures of phosphodiesterase 10 binding with adenosine 3',5'-monophosphate and guanosine 3',5'-monophosphate by hybrid quantum mechanical/molecular mechanical calculations
Abstract
Quantum mechanical/molecular mechanical (QM/MM) geometry optimizations of the X-ray crystal structures of PDE10-AMP (PDB code 2OUN ) and PDE10-GMP (PDB code 2OUQ ) complexes have been performed to characterize the state of the AMP and GMP products, respectively. Results show that only one phosphate oxygen atom (O1) is protonated for both AMP and GMP product complexes. In addition, QM/MM calculations have resolved the orientation of the amide group of Gln726 in PDE10-GMP which was in conflict with the assignment of the guanine group of GMP in the X-ray crystal structure. Calculations reveal that the amide oxygen and nitrogen atom of Gln726 are rotated 180 degrees, resulting in two strong hydrogen bonds formed between the amide group of Gln726 and the guanine group of GMP. Binding free energy calculations for both QM/MM-optimized structures confirm the new conformational assignment of Gln726 in PDE10-GMP. The calculated binding free energy of the rotated structure is approximately 22 kcal/mol lower than the X-ray crystal assignment. The lower energy is mainly derived from the formation of two hydrogen bonds between the amide group of Gln726 and the guanine group of GMP. This implies that the orientation of the amide oxygen and nitrogen atoms in PDE10-AMP is different from PDE10-GMP. Finally, our results help to understand why PDE10 can hydrolyze both cAMP and cGMP.
Figures

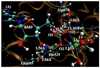




 refers to a coordination bond. There are unfavorable interactions between the GMP guanine group and Gln726 amide group as these two groups are nearly coplanar.
refers to a coordination bond. There are unfavorable interactions between the GMP guanine group and Gln726 amide group as these two groups are nearly coplanar.Similar articles
-
An update view on the substrate recognition mechanism of phosphodiesterases: a computational study of PDE10 and PDE4 bound with cyclic nucleotides.Biopolymers. 2012 Nov;97(11):910-22. doi: 10.1002/bip.22104. Biopolymers. 2012. PMID: 22899366
-
Relative binding free energy calculations of inhibitors to two mutants (Glu46----Ala/Gln) of ribonuclease T1 using molecular dynamics/free energy perturbation approaches.Protein Eng. 1991 Feb;4(3):233-43. doi: 10.1093/protein/4.3.233. Protein Eng. 1991. PMID: 1649996
-
Characterization of a catalytic ligand bridging metal ions in phosphodiesterases 4 and 5 by molecular dynamics simulations and hybrid quantum mechanical/molecular mechanical calculations.Biophys J. 2006 Sep 1;91(5):1858-67. doi: 10.1529/biophysj.106.086835. Biophys J. 2006. PMID: 16912214 Free PMC article.
-
Phosphodiesterase 10 (PDE10) inhibitors: an updated patent review (2014-present).Expert Opin Ther Pat. 2020 Feb;30(2):147-157. doi: 10.1080/13543776.2020.1709444. Epub 2019 Dec 30. Expert Opin Ther Pat. 2020. PMID: 31874060 Review.
-
Crystal structures of some acyclonucleosides with antiviral activity and related compounds.Acta Biochim Pol. 1996;43(1):65-76. Acta Biochim Pol. 1996. PMID: 8790713 Review.
Cited by
-
Microscopic binding of M5 muscarinic acetylcholine receptor with antagonists by homology modeling, molecular docking, and molecular dynamics simulation.J Phys Chem B. 2012 Jan 12;116(1):532-41. doi: 10.1021/jp210579b. Epub 2011 Dec 20. J Phys Chem B. 2012. PMID: 22185605 Free PMC article.
-
Computational determination of binding structures and free energies of phosphodiesterase-2 with benzo[1,4]diazepin-2-one derivatives.J Phys Chem B. 2010 Dec 9;114(48):16020-8. doi: 10.1021/jp1086416. Epub 2010 Nov 15. J Phys Chem B. 2010. PMID: 21077589 Free PMC article.
-
Binding free energies for nicotine analogs inhibiting cytochrome P450 2A6 by a combined use of molecular dynamics simulations and QM/MM-PBSA calculations.Bioorg Med Chem. 2014 Apr 1;22(7):2149-56. doi: 10.1016/j.bmc.2014.02.037. Epub 2014 Mar 3. Bioorg Med Chem. 2014. PMID: 24631364 Free PMC article.
-
Fundamental reaction pathway and free energy profile for hydrolysis of intracellular second messenger adenosine 3',5'-cyclic monophosphate (cAMP) catalyzed by phosphodiesterase-4.J Phys Chem B. 2011 Oct 27;115(42):12208-19. doi: 10.1021/jp205509w. Epub 2011 Oct 5. J Phys Chem B. 2011. PMID: 21973014 Free PMC article.
References
-
- Callahan SM, Cornell NW, Dunlap PV. J. Biol. Chem. 1995;270:17627–17632. - PubMed
-
- Conti M, Jin SLC, Monaco L, Repaske DR, Swinnen JV. Endocr. Rev. 1991;12:218. - PubMed
-
- Houslay MD. Semin. Cell Dev. Biol. 1998;9:161–167. - PubMed
-
- Conti M, Jin SLC. Prog. Nucleic. Acid. Res. 2002;63:1–38. - PubMed
-
- Mehats C, Andersen CB, Filopanti M, Jin SLC, Conti M. Trends Endocrin. Met. 2002;13:29–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

