Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants
- PMID: 20444219
- PMCID: PMC2920365
- DOI: 10.1111/j.1365-3040.2010.02158.x
Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants
Abstract
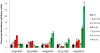
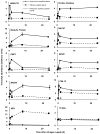
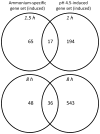
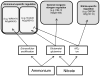
Nitrogen is the only macronutrient that is commonly available to plants in both oxidized and reduced forms, mainly nitrate and ammonium. The physiological and molecular effects of nitrate supply have been well studied, but comparatively little is known about ammonium nutrition and its differential effects on cell function and gene expression. We have used a physiologically realistic hydroponic growth system to compare the transcriptomes and redox status of the roots of ammonium- and nitrate-supplied Arabidopsis thaliana plants. While approximately 60% of nitrogen-regulated genes displayed common responses to both ammonium and nitrate, significant 'nitrate-specific' and 'ammonium-specific' gene sets were identified. Pathways involved in cytokinin response and reductant generation/distribution were specifically altered by nitrate, while a complex biotic stress response and changes in nodulin gene expression were characteristic of ammonium-supplied plants. Nitrate supply was associated with a rapid decrease in H(2)O(2) production, potentially because of an increased export of reductant from the mitochondrial matrix. The underlying basis of the nitrate- and ammonium-specific patterns of gene expression appears to be different signals elaborated from each nitrogen source, including alterations in extracellular pH that are associated with ammonium uptake, downstream metabolites in the ammonium assimilation pathway, and the presence or absence of the nitrate ion.
Figures






References
-
- Beers RF, Sizer IW. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. Journal of Biological Chemistry. 1952;195:133–140. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B. 1995;57:289–300.
-
- Bi YM, Kant S, Clark J, Gidda S, Ming F, Xu J, Rochon A, Shelp BJ, Hao L, Zhao R, Mullen RT, Zhu T, Rothstein SJ. Increased nitrogen use efficiency in transgenic rice plants over-expressing a nitrogen-responsive early nodulin gene identified from rice expression profiling. Plant, Cell and Environment. 2009;32:1749–1760. - PubMed
-
- Bloom AJ. Nitrogen as a limiting factor: Crop acquisition of ammonium and nitrate. In: Jackson LE, editor. Ecology in Agriculture. Academic Press; San Diego, USA: 1997. pp. 145–172.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

