Peripheral blood gene expression patterns discriminate among chronic inflammatory diseases and healthy controls and identify novel targets
- PMID: 20444268
- PMCID: PMC2874757
- DOI: 10.1186/1755-8794-3-15
Peripheral blood gene expression patterns discriminate among chronic inflammatory diseases and healthy controls and identify novel targets
Abstract
Background: Chronic inflammatory diseases including inflammatory bowel disease (IBD; Crohn's disease and ulcerative colitis), psoriasis and rheumatoid arthritis (RA) afflict millions of people worldwide, but their pathogenesis is still not well understood. It is also not well known if distinct changes in gene expression characterize these diseases and if these patterns can discriminate between diseased and control patients and/or stratify the disease. The main focus of our work was the identification of novel markers that overlap among the 3 diseases or discriminate them from each other.
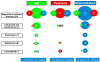
Methods: Diseased (n = 13, n = 15 and n = 12 in IBD, psoriasis and RA respectively) and healthy patients (n = 18) were recruited based on strict inclusion and exclusion criteria; peripheral blood samples were collected by clinicians (30 ml) in Venous Blood Vacuum Collection Tubes containing EDTA and peripheral blood mononuclear cells were separated by Ficoll gradient centrifugation. RNA was extracted using Trizol reagent. Gene expression data was obtained using TaqMan Low Density Array (TLDA) containing 96 genes that were selected by an algorithm and the statistical analyses were performed in Prism by using non-parametric Mann-Whitney U test (P-values < 0.05).
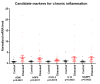
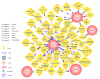
Results: Here we show that using a panel of 96 disease associated genes and measuring mRNA expression levels in peripheral blood derived mononuclear cells; we could identify disease-specific gene panels that separate each disease from healthy controls. In addition, a panel of five genes such as ADM, AQP9, CXCL2, IL10 and NAMPT discriminates between all samples from patients with chronic inflammation and healthy controls. We also found genes that stratify the diseases and separate different subtypes or different states of prognosis in each condition.
Conclusions: These findings and the identification of five universal markers of chronic inflammation suggest that these diseases have a common background in pathomechanism, but still can be separated by peripheral blood gene expression. Importantly, the identified genes can be associated with overlapping biological processes including changed inflammatory response. Gene panels based on such markers can play a major role in the development of personalized medicine, in monitoring disease progression and can lead to the identification of new potential drug targets in chronic inflammation.
Figures



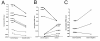
References
-
- Wu F, Dassopoulos T, Cope L, Maitra A, Brant SR, Harris ML, Bayless TM, Parmigiani G, Chakravarti S. Genome-wide gene expression differences in Crohn's disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm Bowel Dis. 2007;13(7):807–821. doi: 10.1002/ibd.20110. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

