An improved method for RNA isolation and cDNA library construction from immature seeds of Jatropha curcas L
- PMID: 20444276
- PMCID: PMC2874567
- DOI: 10.1186/1756-0500-3-126
An improved method for RNA isolation and cDNA library construction from immature seeds of Jatropha curcas L
Abstract
Background: RNA quality and quantity is sometimes unsuitable for cDNA library construction, from plant seeds rich in oil, polysaccharides and other secondary metabolites. Seeds of jatropha (Jatropha curcas L.) are rich in fatty acids/lipids, storage proteins, polysaccharides, and a number of other secondary metabolites that could either bind and/or co-precipitate with RNA, making it unsuitable for downstream applications. Existing RNA isolation methods and commercial kits often fail to deliver high-quality total RNA from immature jatropha seeds for poly(A)+ RNA purification and cDNA synthesis.
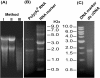
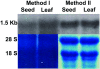
Findings: A protocol has been developed for isolating good quality total RNA from immature jatropha seeds, whereby a combination of the CTAB based RNA extraction method and a silica column of a commercial plant RNA extraction kit is used. The extraction time was reduced from two days to about 3 hours and the RNA was suitable for poly(A)+ RNA purification, cDNA synthesis, cDNA library construction, RT-PCR, and Northern hybridization. Based on sequence information from selected clones and amplified PCR product, the cDNA library seems to be a good source of full-length jatropha genes. The method was equally effective for isolating RNA from mustard and rice seeds.
Conclusions: This is a simple CTAB + silica column method to extract high quality RNA from oil rich immature jatropha seeds that is suitable for several downstream applications. This method takes less time for RNA extraction and is equally effective for other tissues where the quality and quantity of RNA is highly interfered by the presence of fatty acids, polysaccharides and polyphenols.
Figures

Similar articles
-
Isolation of high-quality RNA from various tissues of Jatropha curcas for downstream applications.Anal Biochem. 2011 Jun 1;413(1):63-5. doi: 10.1016/j.ab.2011.01.046. Epub 2011 Mar 5. Anal Biochem. 2011. PMID: 21300020
-
Gene expression profiling identifies pathways involved in seed maturation of Jatropha curcas.BMC Genomics. 2020 Apr 9;21(1):290. doi: 10.1186/s12864-020-6666-1. BMC Genomics. 2020. PMID: 32272887 Free PMC article.
-
Improved and convenient method of RNA isolation from polyphenols and polysaccharide rich plant tissues.Indian J Exp Biol. 2008 Dec;46(12):842-5. Indian J Exp Biol. 2008. PMID: 19245182
-
Potential of Jatropha curcas as a source of renewable oil and animal feed.J Exp Bot. 2009;60(10):2897-905. doi: 10.1093/jxb/erp025. Epub 2009 Feb 13. J Exp Bot. 2009. PMID: 19218317 Review.
-
Current Strategies for the Detoxification of Jatropha curcas Seed Cake: A Review.J Agric Food Chem. 2018 Mar 21;66(11):2510-2522. doi: 10.1021/acs.jafc.7b05691. Epub 2018 Mar 2. J Agric Food Chem. 2018. PMID: 29498277 Review.
Cited by
-
Metabolic engineering of folate and its precursors in Mexican common bean (Phaseolus vulgaris L.).Plant Biotechnol J. 2016 Oct;14(10):2021-32. doi: 10.1111/pbi.12561. Epub 2016 Apr 25. Plant Biotechnol J. 2016. PMID: 26997331 Free PMC article.
-
Dissecting the subcellular membrane proteome reveals enrichment of H+ (co-)transporters and vesicle trafficking proteins in acidic zones of Chara internodal cells.PLoS One. 2018 Aug 29;13(8):e0201480. doi: 10.1371/journal.pone.0201480. eCollection 2018. PLoS One. 2018. PMID: 30157181 Free PMC article.
-
Extractions of High Quality RNA from the Seeds of Jerusalem Artichoke and Other Plant Species with High Levels of Starch and Lipid.Plants (Basel). 2013 Apr 29;2(2):302-16. doi: 10.3390/plants2020302. Plants (Basel). 2013. PMID: 27137377 Free PMC article.
-
RNA-seq data of the Jatropha curcas L. shoot system.Data Brief. 2018 Sep 29;21:71-74. doi: 10.1016/j.dib.2018.09.081. eCollection 2018 Dec. Data Brief. 2018. PMID: 30338276 Free PMC article.
-
An integration of phenotypic and transcriptomic data analysis reveals yield-related hub genes in Jatropha curcas inflorescence.PLoS One. 2018 Sep 21;13(9):e0203441. doi: 10.1371/journal.pone.0203441. eCollection 2018. PLoS One. 2018. PMID: 30240391 Free PMC article.
References
-
- Asif MH, Dhawan P, Nath P. A simple procedure for the isolation of high quality RNA from ripening banana fruit. Plant Mol Biol Rep. 2000;18:109–115. doi: 10.1007/BF02824018. - DOI
-
- Iandolino AB, daSilva FG, Lim H, Choi H, Williams LE, Cook DR. High quality RNA, cDNA, and derived EST libraries from Grapevine (Vitis vinifera L.) Plant Mol Biol Rep. 2004;22:269–278. doi: 10.1007/BF02773137. - DOI
LinkOut - more resources
Full Text Sources

