The structure of adeno-associated virus serotype 3B (AAV-3B): insights into receptor binding and immune evasion
- PMID: 20444480
- PMCID: PMC2885149
- DOI: 10.1016/j.virol.2010.03.027
The structure of adeno-associated virus serotype 3B (AAV-3B): insights into receptor binding and immune evasion
Abstract
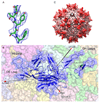
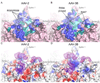
Adeno-associated viruses (AAVs) are leading candidate vectors for human gene therapy. AAV serotypes have broad cellular tropism and use a variety of cellular receptors. AAV serotype 3 binds to heparan sulfate proteoglycan prior to cell entry and is serologically distinct from other serotypes. The capsid features that distinguish AAV-3B from other serotypes are poorly understood. The structure of AAV-3B has been determined to 2.6A resolution from twinned crystals of an infectious virus. The most distinctive structural features are located in regions implicated in receptor and antibody binding, providing insights into the cell entry mechanisms and antigenic nature of AAVs. We show that AAV-3B has a lower affinity for heparin than AAV-2, which can be rationalized by the distinct features of the AAV-3B capsid. The structure of AAV-3B provides an additional foundation for the future engineering of improved gene therapy vectors with modified receptor binding or antigenic characteristics.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures



References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. - PubMed
-
- Agbandje-McKenna M, Llamas-Saiz AL, Wang F, Tattersall P, Rossmann MG. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure. 1998;6(11):1369–1381. - PubMed
-
- Asokan A, Conway JC, Phillips JL, Li C, Hegge J, Sinnott R, Yadav S, DiPrimio N, Nam HJ, Agbandje-McKenna M, McPhee S, Wolff J, Samulski RJ. Reengineering a receptor footprint of adeno-associated virus enables selective and systemic gene transfer to muscle. Nat Biotechnol. 2010;28(1):79–82. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

