Making the message clear: visualizing mRNA localization
- PMID: 20444605
- PMCID: PMC2902723
- DOI: 10.1016/j.tcb.2010.03.006
Making the message clear: visualizing mRNA localization
Abstract
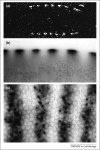
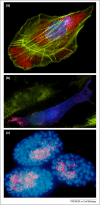
Localized mRNA provides spatial and temporal protein expression essential to cell development and physiology. To explore the mechanisms involved, considerable effort has been spent in establishing new and improved methods for visualizing mRNA. Here, we discuss how these techniques have extended our understanding of intracellular mRNA localization in a variety of organisms. In addition to increased ease and specificity of detection in fixed tissue, in situ hybridization methods now enable examination of mRNA distribution at the ultrastructural level with electron microscopy. Most significantly, methods for following the movement of mRNA in living cells are now in widespread use. These include the introduction of labeled transcripts by microinjection, hybridization based methods using labeled antisense probes and complementary transgenic methods for tagging endogenous mRNAs using bacteriophage components. These technical innovations are now being coupled with super-resolution light microscopy methods and promise to revolutionize our understanding of the dynamics and complexity of the molecular mechanism of mRNA localization.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures



Similar articles
-
Visualizing and Tracking Endogenous mRNAs in Live Drosophila melanogaster Egg Chambers.J Vis Exp. 2019 Jun 4;(148). doi: 10.3791/58545. J Vis Exp. 2019. PMID: 31233020
-
Subcellular localization of mRNA in neuronal cells. Contributions of high-resolution in situ hybridization techniques.Mol Neurobiol. 1998 Dec;18(3):227-46. doi: 10.1007/BF02741301. Mol Neurobiol. 1998. PMID: 10206470 Review.
-
In situ hybridization with non-radioactive digoxigenin-11-UTP-labeled cRNA probes: localization of developmentally regulated mouse tenascin mRNAs.Int J Dev Biol. 1991 Mar;35(1):25-32. Int J Dev Biol. 1991. PMID: 1714291
-
Visualizing mRNAs in fixed and living yeast cells.Methods Mol Biol. 2011;714:203-19. doi: 10.1007/978-1-61779-005-8_13. Methods Mol Biol. 2011. PMID: 21431743
-
Electron microscopy and in situ hybridization: Expression of P2Y2 receptor mRNA in the cerebellum.Methods Mol Biol. 2006;326:151-62. doi: 10.1385/1-59745-007-3:151. Methods Mol Biol. 2006. PMID: 16780199 Review.
Cited by
-
Mining Functional Elements in Messenger RNAs: Overview, Challenges, and Perspectives.Front Plant Sci. 2011 Nov 30;2:84. doi: 10.3389/fpls.2011.00084. eCollection 2011. Front Plant Sci. 2011. PMID: 22639614 Free PMC article.
-
Methods for studying oogenesis.Methods. 2014 Jun 15;68(1):207-17. doi: 10.1016/j.ymeth.2014.01.005. Epub 2014 Jan 17. Methods. 2014. PMID: 24440745 Free PMC article. Review.
-
Condensate functionalization with microtubule motors directs their nucleation in space and allows manipulating RNA localization.EMBO J. 2023 Oct 16;42(20):e114106. doi: 10.15252/embj.2023114106. Epub 2023 Sep 19. EMBO J. 2023. PMID: 37724036 Free PMC article.
-
Fluorescent turn-on probes for wash-free mRNA imaging via covalent site-specific enzymatic labeling.Chem Sci. 2017 Oct 1;8(10):7169-7173. doi: 10.1039/c7sc03150e. Epub 2017 Aug 29. Chem Sci. 2017. PMID: 29081948 Free PMC article.
-
How the sea squirt nucleus tells mesoderm not to be endoderm.Dev Cell. 2010 Oct 19;19(4):487-8. doi: 10.1016/j.devcel.2010.10.002. Dev Cell. 2010. PMID: 20951340 Free PMC article.
References
-
- Lopez de Heredia M., Jansen R.P. mRNA localization and the cytoskeleton. Curr. Opin. Cell Biol. 2004;16:80–85. - PubMed
-
- St Johnston D. Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 2005;6:363–375. - PubMed
-
- Levsky J.M., Singer R.H. Fluorescence in situ hybridization: past, present and future. J. Cell. Sci. 2003;116:2833–2838. - PubMed
-
- Tyagi S. Imaging intracellular RNA distribution and dynamics in living cells. Nat. Methods. 2009;6:331–338. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

