Methylation of ribosomal protein L42 regulates ribosomal function and stress-adapted cell growth
- PMID: 20444689
- PMCID: PMC2903366
- DOI: 10.1074/jbc.M110.132274
Methylation of ribosomal protein L42 regulates ribosomal function and stress-adapted cell growth
Abstract
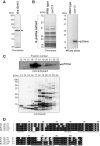
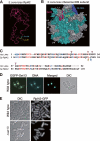
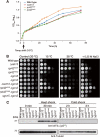
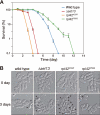
Lysine methylation is one of the most common protein modifications. Although lysine methylation of histones has been extensively studied and linked to gene regulation, that of non-histone proteins remains incompletely understood. Here, we show a novel regulatory role of ribosomal protein methylation. Using an in vitro methyltransferase assay, we found that Schizosaccharomyces pombe Set13, a SET domain protein encoded by SPAC688.14, specifically methylates lysine 55 of ribosomal protein L42 (Rpl42). Mass spectrometric analysis revealed that endogenous Rpl42 is monomethylated at lysine 55 in wild-type S. pombe cells and that the methylation is lost in Delta set13 mutant cells. Delta set13 and Rpl42 methylation-deficient mutant S. pombe cells showed higher cycloheximide sensitivity and defects in stress-responsive growth control compared with wild type. Genetic analyses suggested that the abnormal growth phenotype was distinct from the conserved stress-responsive pathway that modulates translation initiation. Furthermore, the Rpl42 methylation-deficient mutant cells showed a reduced ability to survive after entering stationary phase. These results suggest that Rpl42 methylation plays direct roles in ribosomal function and cell proliferation control independently of the general stress-response pathway.
Figures




Similar articles
-
A conserved SET domain methyltransferase, Set11, modifies ribosomal protein Rpl12 in fission yeast.J Biol Chem. 2008 Mar 14;283(11):7185-95. doi: 10.1074/jbc.M709429200. Epub 2008 Jan 14. J Biol Chem. 2008. PMID: 18195021
-
PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits.EMBO J. 2004 Jul 7;23(13):2641-50. doi: 10.1038/sj.emboj.7600265. Epub 2004 Jun 3. EMBO J. 2004. PMID: 15175657 Free PMC article.
-
Ribosomal protein eL42 contributes to the catalytic activity of the yeast ribosome at the elongation step of translation.Biochimie. 2019 Mar;158:20-33. doi: 10.1016/j.biochi.2018.12.005. Epub 2018 Dec 11. Biochimie. 2019. PMID: 30550856
-
[Ash2, a subunit of histone H3K4 methyltransferase complex, is involved in the sporulation in Schizosaccharomyces pombe].Yi Chuan. 2014 Sep;36(9):943-51. doi: 10.3724/SP.J.1005.2014.0943. Yi Chuan. 2014. PMID: 25252312 Chinese.
-
Research Progress of Ribosomal Proteins in Reproductive Development.Int J Mol Sci. 2024 Dec 7;25(23):13151. doi: 10.3390/ijms252313151. Int J Mol Sci. 2024. PMID: 39684863 Free PMC article. Review.
Cited by
-
A Cloning-Free Method for CRISPR/Cas9-Mediated Genome Editing in Fission Yeast.G3 (Bethesda). 2018 May 31;8(6):2067-2077. doi: 10.1534/g3.118.200164. G3 (Bethesda). 2018. PMID: 29703785 Free PMC article.
-
Regulation of the SUV39H Family Methyltransferases: Insights from Fission Yeast.Biomolecules. 2023 Mar 25;13(4):593. doi: 10.3390/biom13040593. Biomolecules. 2023. PMID: 37189341 Free PMC article. Review.
-
S-Adenosylmethionine Synthetase Is Required for Cell Growth, Maintenance of G0 Phase, and Termination of Quiescence in Fission Yeast.iScience. 2018 Jul 27;5:38-51. doi: 10.1016/j.isci.2018.06.011. Epub 2018 Jun 30. iScience. 2018. PMID: 30240645 Free PMC article.
-
Ribosome-associated proteins: unwRAPping ribosome heterogeneity in the twenty-first century.Philos Trans R Soc Lond B Biol Sci. 2025 Mar 6;380(1921):20230378. doi: 10.1098/rstb.2023.0378. Epub 2025 Mar 6. Philos Trans R Soc Lond B Biol Sci. 2025. PMID: 40045784 Review.
-
Structural basis for the inhibition of the eukaryotic ribosome.Nature. 2014 Sep 25;513(7519):517-22. doi: 10.1038/nature13737. Epub 2014 Sep 10. Nature. 2014. PMID: 25209664
References
-
- Clarke S. G., Tamanoi F. (eds) (2006) The Enzymes, 3rd Ed., Vol. 24, Academic Press, San Diego
-
- Lachner M., Jenuwein T. (2002) Curr. Opin. Cell Biol. 14, 286–298 - PubMed
-
- Martin C., Zhang Y. (2005) Nat. Rev. Mol. Cell Biol. 6, 838–849 - PubMed
-
- Klose R. J., Zhang Y. (2007) Nat. Rev. Mol. Cell Biol. 8, 307–318 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

