LPA19, a Psb27 homolog in Arabidopsis thaliana, facilitates D1 protein precursor processing during PSII biogenesis
- PMID: 20444695
- PMCID: PMC2898400
- DOI: 10.1074/jbc.M110.105064
LPA19, a Psb27 homolog in Arabidopsis thaliana, facilitates D1 protein precursor processing during PSII biogenesis
Abstract
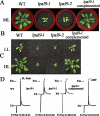
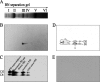
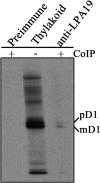
The biogenesis and assembly of photosystem II (PSII) are mainly regulated by the nuclear-encoded factors. To further identify the novel components involved in PSII biogenesis, we isolated and characterized a high chlorophyll fluorescence low psii accumulation19 (lpa19) mutant, which is defective in PSII biogenesis. LPA19 encodes a Psb27 homolog (At1g05385). Interestingly, another Psb27 homolog (At1g03600) in Arabidopsis was revealed to be required for the efficient repair of photodamaged PSII. These results suggest that the Psb27 homologs play distinct functions in PSII biogenesis and repair in Arabidopsis. Chloroplast protein labeling assays showed that the C-terminal processing of D1 in the lpa19 mutant was impaired. Protein overlay assays provided evidence that LPA19 interacts with D1, and coimmunoprecipitation analysis demonstrated that LPA19 interacts with mature D1 (mD1) and precursor D1 (pD1). Moreover, LPA19 protein was shown to specifically interact with the soluble C terminus present in the precursor and mature D1 through yeast two-hybrid analyses. Thus, these studies suggest that LPA19 is involved in facilitating the D1 precursor protein processing in Arabidopsis.
Figures





Similar articles
-
A Psb27 homologue in Arabidopsis thaliana is required for efficient repair of photodamaged photosystem II.Plant Mol Biol. 2006 Jul;61(4-5):567-75. doi: 10.1007/s11103-006-0031-x. Plant Mol Biol. 2006. PMID: 16897475
-
The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly.Plant Cell. 2010 Oct;22(10):3439-60. doi: 10.1105/tpc.110.077453. Epub 2010 Oct 5. Plant Cell. 2010. PMID: 20923938 Free PMC article.
-
LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana.Plant Cell. 2007 Jun;19(6):1980-93. doi: 10.1105/tpc.107.050526. Epub 2007 Jun 29. Plant Cell. 2007. Retraction in: Plant Cell. 2016 Dec;28(12):3061. doi: 10.1105/tpc.16.00881. PMID: 17601825 Free PMC article. Retracted.
-
Revisiting the photosystem II repair cycle.Plant Signal Behav. 2016 Sep;11(9):e1218587. doi: 10.1080/15592324.2016.1218587. Plant Signal Behav. 2016. PMID: 27494214 Free PMC article. Review.
-
Processing of D1 Protein: A Mysterious Process Carried Out in Thylakoid Lumen.Int J Mol Sci. 2022 Feb 25;23(5):2520. doi: 10.3390/ijms23052520. Int J Mol Sci. 2022. PMID: 35269663 Free PMC article. Review.
Cited by
-
How to build functional thylakoid membranes: from plastid transcription to protein complex assembly.Planta. 2013 Feb;237(2):413-28. doi: 10.1007/s00425-012-1752-5. Epub 2012 Sep 14. Planta. 2013. PMID: 22976450 Free PMC article. Review.
-
Structural, functional and auxiliary proteins of photosystem II.Photosynth Res. 2013 Oct;116(2-3):167-88. doi: 10.1007/s11120-013-9803-8. Epub 2013 Feb 17. Photosynth Res. 2013. PMID: 23417641 Review.
-
PsbN is required for assembly of the photosystem II reaction center in Nicotiana tabacum.Plant Cell. 2014 Mar;26(3):1183-99. doi: 10.1105/tpc.113.120444. Epub 2014 Mar 11. Plant Cell. 2014. PMID: 24619613 Free PMC article.
-
Analysis of essential Arabidopsis nuclear genes encoding plastid-targeted proteins.PLoS One. 2013 Sep 4;8(9):e73291. doi: 10.1371/journal.pone.0073291. eCollection 2013. PLoS One. 2013. PMID: 24023856 Free PMC article.
-
Structural insights into a dimeric Psb27-photosystem II complex from a cyanobacterium Thermosynechococcus vulcanus.Proc Natl Acad Sci U S A. 2021 Feb 2;118(5):e2018053118. doi: 10.1073/pnas.2018053118. Proc Natl Acad Sci U S A. 2021. PMID: 33495333 Free PMC article.
References
-
- Iwata S., Barber J. (2004) Curr. Opin. Plant. Biol. 14, 447–453 - PubMed
-
- Nelson N., Yocum C. F. (2006) Annu. Rev. Plant Biol. 57, 521–565 - PubMed
-
- Ferreira K. N., Iverson T. M., Maghlaoui K., Barber J., Iwata S. (2004) Science 303, 1831–1838 - PubMed
-
- Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. (2005) Nature 438, 1040–1044 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

