Modulation of UvrD helicase activity by covalent DNA-protein cross-links
- PMID: 20444702
- PMCID: PMC2898412
- DOI: 10.1074/jbc.M109.078964
Modulation of UvrD helicase activity by covalent DNA-protein cross-links
Abstract
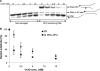
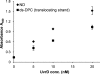
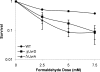
UvrD (DNA helicase II) has been implicated in DNA replication, DNA recombination, nucleotide excision repair, and methyl-directed mismatch repair. The enzymatic function of UvrD is to translocate along a DNA strand in a 3' to 5' direction and unwind duplex DNA utilizing a DNA-dependent ATPase activity. In addition, UvrD interacts with many other proteins involved in the above processes and is hypothesized to facilitate protein turnover, thus promoting further DNA processing. Although UvrD interactions with proteins bound to DNA have significant biological implications, the effects of covalent DNA-protein cross-links on UvrD helicase activity have not been characterized. Herein, we demonstrate that UvrD-catalyzed strand separation was inhibited on a DNA strand to which a 16-kDa protein was covalently bound. Our sequestration studies suggest that the inhibition of UvrD activity is most likely due to a translocation block and not helicase sequestration on the cross-link-containing DNA substrate. In contrast, no inhibition of UvrD-catalyzed strand separation was apparent when the protein was linked to the complementary strand. The latter result is surprising given the earlier observations that the DNA in this covalent complex is severely bent ( approximately 70 degrees ), with both DNA strands making multiple contacts with the cross-linked protein. In addition, UvrD was shown to be required for replication of plasmid DNAs containing covalent DNA-protein complexes. Combined, these data suggest a critical role for UvrD in the processing of DNA-protein cross-links.
Figures




Similar articles
-
The UvrD helicase and its modulation by the mismatch repair protein MutL.Nucleic Acids Res. 2006;34(15):4089-97. doi: 10.1093/nar/gkl450. Epub 2006 Aug 25. Nucleic Acids Res. 2006. PMID: 16935885 Free PMC article.
-
Large domain movements upon UvrD dimerization and helicase activation.Proc Natl Acad Sci U S A. 2017 Nov 14;114(46):12178-12183. doi: 10.1073/pnas.1712882114. Epub 2017 Oct 30. Proc Natl Acad Sci U S A. 2017. PMID: 29087333 Free PMC article.
-
Stimulation of UvrD helicase by UvrAB.J Biol Chem. 2009 Apr 3;284(14):9612-23. doi: 10.1074/jbc.M808030200. Epub 2009 Feb 10. J Biol Chem. 2009. PMID: 19208629 Free PMC article.
-
lon incompatibility associated with mutations causing SOS induction: null uvrD alleles induce an SOS response in Escherichia coli.J Bacteriol. 2000 Jun;182(11):3151-7. doi: 10.1128/JB.182.11.3151-3157.2000. J Bacteriol. 2000. PMID: 10809694 Free PMC article.
-
UvrD helicase: an old dog with a new trick: how one step backward leads to many steps forward.Bioessays. 2015 Jan;37(1):12-9. doi: 10.1002/bies.201400106. Epub 2014 Oct 27. Bioessays. 2015. PMID: 25345862 Free PMC article. Review.
Cited by
-
Formation and repair of DNA-protein crosslink damage.Sci China Life Sci. 2017 Oct;60(10):1065-1076. doi: 10.1007/s11427-017-9183-4. Epub 2017 Oct 30. Sci China Life Sci. 2017. PMID: 29098631 Free PMC article. Review.
-
Functions that protect Escherichia coli from DNA-protein crosslinks.DNA Repair (Amst). 2015 Apr;28:48-59. doi: 10.1016/j.dnarep.2015.01.016. Epub 2015 Feb 7. DNA Repair (Amst). 2015. PMID: 25731940 Free PMC article.
-
Close encounters for the first time: Helicase interactions with DNA damage.DNA Repair (Amst). 2015 Sep;33:43-59. doi: 10.1016/j.dnarep.2015.06.003. Epub 2015 Jun 16. DNA Repair (Amst). 2015. PMID: 26160335 Free PMC article. Review.
-
Detection of DNA-protein crosslinks (DPCs) by novel direct fluorescence labeling methods: distinct stabilities of aldehyde and radiation-induced DPCs.Nucleic Acids Res. 2012 Oct;40(18):e143. doi: 10.1093/nar/gks601. Epub 2012 Jun 22. Nucleic Acids Res. 2012. PMID: 22730301 Free PMC article.
-
S-phase sensing of DNA-protein crosslinks triggers TopBP1-independent ATR activation and p53-mediated cell death by formaldehyde.Cell Cycle. 2012 Jul 1;11(13):2526-37. doi: 10.4161/cc.20905. Epub 2012 Jul 1. Cell Cycle. 2012. PMID: 22722496 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

