Dissecting the in vivo metabolic potential of two human gut acetogens
- PMID: 20444704
- PMCID: PMC2903421
- DOI: 10.1074/jbc.M110.117713
Dissecting the in vivo metabolic potential of two human gut acetogens
Abstract
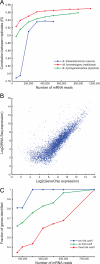
Fermenting microbial communities generate hydrogen; its removal through the production of acetate, methane, or hydrogen sulfide modulates the efficiency of energy extraction from available nutrients in many ecosystems. We noted that pathway components for acetogenesis are more abundantly and consistently represented in the gut microbiomes of monozygotic twins and their mothers than components for methanogenesis or sulfate reduction and subsequently analyzed the metabolic potential of two sequenced human gut acetogens, Blautia hydrogenotrophica and Marvinbryantia formatexigens in vitro and in the intestines of gnotobiotic mice harboring a prominent saccharolytic bacterium. To do so, we developed a generally applicable method for multiplex sequencing of expressed microbial mRNAs (microbial RNA-Seq) and, together with mass spectrometry of metabolites, showed that these organisms have distinct patterns of substrate utilization. B. hydrogenotrophica targets aliphatic and aromatic amino acids. It increases the efficiency of fermentation by consuming reducing equivalents, thereby maintaining a high NAD(+)/NADH ratio and boosting acetate production. In contrast, M. formatexigens consumes oligosaccharides, does not impact the redox state of the gut, and boosts the yield of succinate. These findings have strategic implications for those who wish to manipulate the hydrogen economy of gut microbial communities in ways that modulate energy harvest.
Figures

References
-
- Flint H. J., Bayer E. A., Rincon M. T., Lamed R., White B. A. (2008) Nat. Rev. Microbiol. 6, 121–131 - PubMed
-
- Macfarlane S., Macfarlane G. T. (2003) Proc. Nutr. Soc. 62, 67–72 - PubMed
-
- Wolin M. J., Miller T. L. (1983) Fed. Proc. 42, 109–113 - PubMed
-
- McNeil N. I. (1984) Am. J. Clin. Nutr. 39, 338–342 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

