GREBP, a cGMP-response element-binding protein repressing the transcription of natriuretic peptide receptor 1 (NPR1/GCA)
- PMID: 20444705
- PMCID: PMC2898340
- DOI: 10.1074/jbc.M109.061622
GREBP, a cGMP-response element-binding protein repressing the transcription of natriuretic peptide receptor 1 (NPR1/GCA)
Abstract
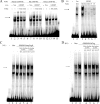
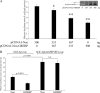
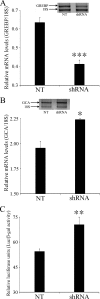
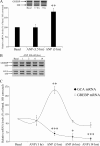
NPR1/GCA (natriuretic peptide receptor 1/guanylyl cyclase A) expression is controlled by several agents, including ANP (atrial natriuretic peptide). After ANP stimulation, NPR1/GCA down-regulates the transcriptional activity of its gene via a cGMP-dependent mechanism. Because we previously identified a cis-acting element responsible for this cGMP sensitivity, we proceed here to explore novel putative protein binding to cGMP-response element (cGMP-RE). Using the yeast one-hybrid technique with a human kidney cDNA library, we identified a strong positive clone able to bind cGMP-RE. The clone was derived from 1083-bp-long cDNA of a gene of yet unknown function localized on human chromosome 1 (1p33.36). We named this new protein GREBP (for cGMP-response element-binding protein). DNA binding assays showed 18-fold higher cGMP-RE binding capacity than the controls, whereas an electromobility shift assay indicated a specific binding for the cGMP-RE, and chromatin immunoprecipitation confirmed the binding of GREBP to the element under physiological conditions. By acting on cGMP-RE, GREBP inhibited the expression of a luciferase-coupled NPR1 promoter construct. In H295R cells, ANP heightened GREBP expression by 60% after just 3 h of treatment while inhibiting NPR1/GCA expression by 30%. Silencing GREBP with specific small interfering RNA increased the activity of the luciferase-coupled NPR1 promoter and GCA/NPR1 mRNA levels. GREBP is a nuclear protein mainly expressed in the heart. We report here the existence of a human-specific gene that acts as a transcriptional repressor of the NPR1/GCA gene.
Figures




Similar articles
-
Characterization of a cGMP-response element in the guanylyl cyclase/natriuretic peptide receptor A gene promoter.Hypertension. 2004 Jun;43(6):1270-8. doi: 10.1161/01.HYP.0000126920.93207.53. Epub 2004 Apr 19. Hypertension. 2004. PMID: 15096467
-
Valporic acid enhances the Atrial Natriuretic Peptide (ANP) mediated anti-hypertrophic activity by modulating the Npr1 gene transcription in H9c2 cells in vitro.Eur J Pharmacol. 2017 Oct 15;813:94-104. doi: 10.1016/j.ejphar.2017.07.042. Epub 2017 Jul 22. Eur J Pharmacol. 2017. PMID: 28743391
-
Hypertension associated with decreased testosterone levels in natriuretic peptide receptor-A gene-knockout and gene-duplicated mutant mouse models.Endocrinology. 1999 Nov;140(11):5112-9. doi: 10.1210/endo.140.11.7121. Endocrinology. 1999. PMID: 10537139
-
Regulation of guanylyl cyclase/natriuretic peptide receptor-A gene expression.Peptides. 2005 Jun;26(6):1009-23. doi: 10.1016/j.peptides.2004.09.022. Epub 2005 Apr 13. Peptides. 2005. PMID: 15911069 Review.
-
Molecular and genetic aspects of guanylyl cyclase natriuretic peptide receptor-A in regulation of blood pressure and renal function.Physiol Genomics. 2018 Nov 1;50(11):913-928. doi: 10.1152/physiolgenomics.00083.2018. Epub 2018 Aug 31. Physiol Genomics. 2018. PMID: 30169131 Free PMC article. Review.
Cited by
-
Association of natriuretic peptides and receptor activity with cardio-metabolic health at middle age.Sci Rep. 2024 Apr 30;14(1):9919. doi: 10.1038/s41598-024-60677-4. Sci Rep. 2024. PMID: 38689031 Free PMC article.
-
ANP-induced signaling cascade and its implications in renal pathophysiology.Am J Physiol Renal Physiol. 2015 May 15;308(10):F1047-55. doi: 10.1152/ajprenal.00164.2014. Epub 2015 Jan 28. Am J Physiol Renal Physiol. 2015. PMID: 25651559 Free PMC article. Review.
-
Binding of activating transcription factor 6 to the A5/Core of the rat insulin II gene promoter does not mediate its transcriptional repression.J Mol Endocrinol. 2011 Sep 30;47(3):273-83. doi: 10.1530/JME-11-0016. Print 2011 Dec. J Mol Endocrinol. 2011. PMID: 21821716 Free PMC article.
-
Recovery of rhythmic activity in a central pattern generator: analysis of the role of neuromodulator and activity-dependent mechanisms.J Comput Neurosci. 2011 Nov;31(3):685-99. doi: 10.1007/s10827-011-0338-8. Epub 2011 May 15. J Comput Neurosci. 2011. PMID: 21573963 Free PMC article.
-
Histone deacetylase inhibitors modulate the transcriptional regulation of guanylyl cyclase/natriuretic peptide receptor-a gene: interactive roles of modified histones, histone acetyltransferase, p300, AND Sp1.J Biol Chem. 2014 Mar 7;289(10):6991-7002. doi: 10.1074/jbc.M113.511444. Epub 2014 Jan 22. J Biol Chem. 2014. PMID: 24451378 Free PMC article.
References
-
- de Bold A. J., Borenstein H. B., Veress A. T., Sonnenberg H. (1981) Life Sci. 28, 89–94 - PubMed
-
- Hamet P., Tremblay J., Pang S. C., Garcia R., Thibault G., Gutkowska J., Cantin M., Genest J. (1984) Biochem. Biophys. Res. Commun. 123, 515–527 - PubMed
-
- Tremblay J., Gerzer R., Pang S. C., Cantin M., Genest J., Hamet P. (1986) FEBS Lett. 194, 210–214 - PubMed
-
- Inagami T. (1989) J. Biol. Chem. 264, 3043–3046 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

