MetExplore: a web server to link metabolomic experiments and genome-scale metabolic networks
- PMID: 20444866
- PMCID: PMC2896158
- DOI: 10.1093/nar/gkq312
MetExplore: a web server to link metabolomic experiments and genome-scale metabolic networks
Abstract
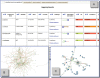
High-throughput metabolomic experiments aim at identifying and ultimately quantifying all metabolites present in biological systems. The metabolites are interconnected through metabolic reactions, generally grouped into metabolic pathways. Classical metabolic maps provide a relational context to help interpret metabolomics experiments and a wide range of tools have been developed to help place metabolites within metabolic pathways. However, the representation of metabolites within separate disconnected pathways overlooks most of the connectivity of the metabolome. By definition, reference pathways cannot integrate novel pathways nor show relationships between metabolites that may be linked by common neighbours without being considered as joint members of a classical biochemical pathway. MetExplore is a web server that offers the possibility to link metabolites identified in untargeted metabolomics experiments within the context of genome-scale reconstructed metabolic networks. The analysis pipeline comprises mapping metabolomics data onto the specific metabolic network of an organism, then applying graph-based methods and advanced visualization tools to enhance data analysis. The MetExplore web server is freely accessible at http://metexplore.toulouse.inra.fr.
Figures
References
-
- Francke C, Siezen RJ, Teusink B. Reconstructing the metabolic network of a bacterium from its genome. Trends Microbiol. 2005;13:550–558. - PubMed
-
- Lacroix V, Cottret L, Thébault P, Sagot M.-F. An introduction to metabolic networks and their structural analysis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2008;5:594–617. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases