Frog2: Efficient 3D conformation ensemble generator for small compounds
- PMID: 20444874
- PMCID: PMC2896087
- DOI: 10.1093/nar/gkq325
Frog2: Efficient 3D conformation ensemble generator for small compounds
Abstract
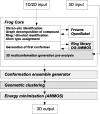
Frog is a web tool dedicated to small compound 3D generation. Here we present the new version, Frog2, which allows the generation of conformation ensembles of small molecules starting from either 1D, 2D or 3D description of the compounds. From a compound description in one of the SMILES, SDF or mol2 formats, the server will return an ensemble of diverse conformers generated using a two stage Monte Carlo approach in the dihedral space. When starting from 1D or 2D description of compounds, Frog2 is capable to detect the sites of ambiguous stereoisomery, and thus to sample different stereoisomers. Frog2 also embeds new energy minimization and ring generation facilities that solve the problem of some missing cycle structures in the Frog1 ring library. Finally, the optimized generator of conformation ensembles in Frog2 results in a gain of computational time permitting Frog2 to be up to 20 times faster that Frog1, while producing satisfactory conformations in terms of structural quality and conformational diversity. The high speed and the good quality of generated conformational ensembles makes it possible the treatment of larger compound collections using Frog2. The server and documentation are freely available at http://bioserv.rpbs.univ-paris-diderot.fr/Frog2.
Figures

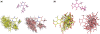
Similar articles
-
The FAF-Drugs2 server: a multistep engine to prepare electronic chemical compound collections.Bioinformatics. 2011 Jul 15;27(14):2018-20. doi: 10.1093/bioinformatics/btr333. Epub 2011 Jun 2. Bioinformatics. 2011. PMID: 21636592
-
Frog: a FRee Online druG 3D conformation generator.Nucleic Acids Res. 2007 Jul;35(Web Server issue):W568-72. doi: 10.1093/nar/gkm289. Epub 2007 May 7. Nucleic Acids Res. 2007. PMID: 17485475 Free PMC article.
-
MTiOpenScreen: a web server for structure-based virtual screening.Nucleic Acids Res. 2015 Jul 1;43(W1):W448-54. doi: 10.1093/nar/gkv306. Epub 2015 Apr 8. Nucleic Acids Res. 2015. PMID: 25855812 Free PMC article.
-
Recent developments of the chemistry development kit (CDK) - an open-source java library for chemo- and bioinformatics.Curr Pharm Des. 2006;12(17):2111-20. doi: 10.2174/138161206777585274. Curr Pharm Des. 2006. PMID: 16796559 Review.
-
Conformational sampling and energetics of drug-like molecules.Curr Med Chem. 2009;16(26):3381-413. doi: 10.2174/092986709789057680. Epub 2009 Sep 1. Curr Med Chem. 2009. PMID: 19515013 Review.
Cited by
-
Open source molecular modeling.J Mol Graph Model. 2016 Sep;69:127-43. doi: 10.1016/j.jmgm.2016.07.008. Epub 2016 Jul 30. J Mol Graph Model. 2016. PMID: 27631126 Free PMC article. Review.
-
Best-Practice DFT Protocols for Basic Molecular Computational Chemistry.Angew Chem Int Ed Engl. 2022 Oct 17;61(42):e202205735. doi: 10.1002/anie.202205735. Epub 2022 Sep 14. Angew Chem Int Ed Engl. 2022. PMID: 36103607 Free PMC article. Review.
-
Flexibility and binding affinity in protein-ligand, protein-protein and multi-component protein interactions: limitations of current computational approaches.J R Soc Interface. 2012 Jan 7;9(66):20-33. doi: 10.1098/rsif.2011.0584. Epub 2011 Oct 12. J R Soc Interface. 2012. PMID: 21993006 Free PMC article. Review.
-
A UPLC-DAD-Based Bio-Screening Assay for the Evaluation of the Angiotensin Converting Enzyme Inhibitory Potential of Plant Extracts and Compounds: Pyrroquinazoline Alkaloids from Adhatoda vasica as a Case Study.Molecules. 2021 Nov 18;26(22):6971. doi: 10.3390/molecules26226971. Molecules. 2021. PMID: 34834066 Free PMC article.
-
Tyrosine kinase syk non-enzymatic inhibitors and potential anti-allergic drug-like compounds discovered by virtual and in vitro screening.PLoS One. 2011;6(6):e21117. doi: 10.1371/journal.pone.0021117. Epub 2011 Jun 20. PLoS One. 2011. PMID: 21701581 Free PMC article.
References
-
- Clark D. What has virtual screening ever done for drug discovery? Expert. Opin. Drug. Discov. 2008;3:841–851. - PubMed
-
- Douguet D. Ligand-based approaches in virtual screening. Cur. Comput. Aided Drug Des. 2008;4:180–190.
-
- Pajeva IK, Globisch C, Wiese M. Combined pharmacophore modeling, docking, and 3D QSAR studies of ABCB1 and ABCC1 transporter inhibitors. ChemMedChem. 2009;4:1883–1896. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

