Decoding accuracy in eRF1 mutants and its correlation with pleiotropic quantitative traits in yeast
- PMID: 20444877
- PMCID: PMC2938225
- DOI: 10.1093/nar/gkq338
Decoding accuracy in eRF1 mutants and its correlation with pleiotropic quantitative traits in yeast
Abstract
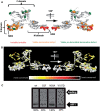
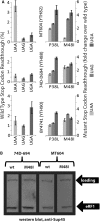
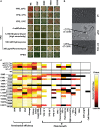
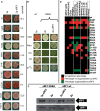
Translation termination in eukaryotes typically requires the decoding of one of three stop codons UAA, UAG or UGA by the eukaryotic release factor eRF1. The molecular mechanisms that allow eRF1 to decode either A or G in the second nucleotide, but to exclude UGG as a stop codon, are currently not well understood. Several models of stop codon recognition have been developed on the basis of evidence from mutagenesis studies, as well as studies on the evolutionary sequence conservation of eRF1. We show here that point mutants of Saccharomyces cerevisiae eRF1 display significant variability in their stop codon read-through phenotypes depending on the background genotype of the strain used, and that evolutionary conservation of amino acids in eRF1 is only a poor indicator of the functional importance of individual residues in translation termination. We further show that many phenotypes associated with eRF1 mutants are quantitatively unlinked with translation termination defects, suggesting that the evolutionary history of eRF1 was shaped by a complex set of molecular functions in addition to translation termination. We reassess current models of stop-codon recognition by eRF1 in the light of these new data.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

