tRNASec is transcribed by RNA polymerase II in Trypanosoma brucei but not in humans
- PMID: 20444878
- PMCID: PMC2943599
- DOI: 10.1093/nar/gkq345
tRNASec is transcribed by RNA polymerase II in Trypanosoma brucei but not in humans
Abstract
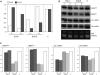
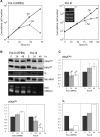
Nuclear-encoded tRNAs are universally transcribed by RNA polymerase III (Pol-III) and contain intragenic promoters. Transcription of vertebrate tRNA(Sec) however requires extragenic promoters similar to Pol-III transcribed U6 snRNA. Here, we present a comparative analysis of tRNA(Sec) transcription in humans and the parasitic protozoa Trypanosoma brucei, two evolutionary highly diverged eukaryotes. RNAi-mediated ablation of Pol-II and Pol-III as well as oligo-dT induced transcription termination show that the human tRNA(Sec) is a Pol-III transcript. In T. brucei protein-coding genes are polycistronically transcribed by Pol-II and processed by trans-splicing and polyadenylation. tRNA genes are generally clustered in between polycistrons. However, the trypanosomal tRNA(Sec) genes are embedded within a polycistron. Their transcription is sensitive to α-amanitin and RNAi-mediated ablation of Pol-II, but not of Pol-III. Ectopic expression of the tRNA(Sec) outside but not inside a polycistron requires an added external promoter. These experiments demonstrate that trypanosomal tRNA(Sec), in contrast to its human counterpart, is transcribed by Pol-II. Synteny analysis shows that in trypanosomatids the tRNA(Sec) gene can be found in two different polycistrons, suggesting that it has evolved twice independently. Moreover, intron-encoded tRNAs are present in a number of eukaryotic genomes indicating that Pol-II transcription of tRNAs may not be restricted to trypanosomatids.
Figures

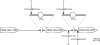


References
-
- Commans S, Böck A. Selenocysteine inserting tRNAs: an overview. FEMS Microbiol. Rev. 1999;23:335–351. - PubMed
-
- Hatfield DL, Carlson BA, Xu XM, Mix H, Gladyshev VN. Selenocysteine incorporation machinery and the role of selenoproteins in development and health. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:97–142. - PubMed
-
- Lee BJ, Kang SG, Hatfield D. Transcription of Xenopus selenocysteine tRNA Ser (formerly designated opal suppressor phosphoserine tRNA) gene is directed by multiple 5′-extragenic regulatory elements. J. Biol. Chem. 1989;264:9696–9702. - PubMed

