Intra- and interhost evolutionary dynamics of equine influenza virus
- PMID: 20444896
- PMCID: PMC2898244
- DOI: 10.1128/JVI.00112-10
Intra- and interhost evolutionary dynamics of equine influenza virus
Abstract
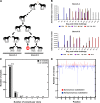
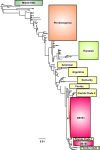
Determining the evolutionary basis of cross-species transmission and immune evasion is key to understanding the mechanisms that control the emergence of either new viruses or novel antigenic variants with pandemic potential. The hemagglutinin glycoprotein of influenza A viruses is a critical host range determinant and a major target of neutralizing antibodies. Equine influenza virus (EIV) is a significant pathogen of the horse that causes periodical outbreaks of disease even in populations with high vaccination coverage. EIV has also jumped the species barrier and emerged as a novel respiratory pathogen in dogs, canine influenza virus. We studied the dynamics of equine influenza virus evolution in horses at the intrahost level and how this evolutionary process is affected by interhost transmission in a natural setting. To this end, we performed clonal sequencing of the hemagglutinin 1 gene derived from individual animals at different times postinfection. Our results show that despite the population consensus sequence remaining invariant, genetically distinct subpopulations persist during the course of infection and are also transmitted, with some variants likely to change antigenicity. We also detected a natural case of mixed infection in an animal infected during an outbreak of equine influenza, raising the possibility of reassortment between different strains of virus. In sum, our data suggest that transmission bottlenecks may not be as narrow as originally perceived and that the genetic diversity required to adapt to new host species may be partially present in the donor host and potentially transmitted to the recipient host.
Figures


References
-
- Beigel, J. H., J. Farrar, A. M. Han, F. G. Hayden, R. Hyer, M. D. de Jong, S. Lochindarat, T. K. Nguyen, T. H. Nguyen, T. H. Tran, A. Nicoll, S. Touch, and K. Y. Yuen. 2005. Avian influenza A (H5N1) infection in humans. N. Engl. J. Med. 353:1374-1385. - PubMed
-
- Bryant, N. A., A. S. Rash, C. A. Russell, J. Ross, A. Cooke, S. Bowman, S. Macrae, N. S. Lewis, R. Paillot, R. Zanoni, H. Meier, L. A. Griffiths, J. M. Daly, A. Tiwari, T. M. Chambers, J. R. Newton, and D. M. Elton. 2009. Antigenic and genetic variations in European and North American equine influenza virus strains (H3N8) isolated from 2006 to 2007. Vet. Microbiol. 138:41-52. - PubMed
-
- Cook, R. F., R. Sinclair, and J. A. Mumford. 1988. Detection of influenza nucleoprotein antigen in nasal secretions from horses infected with A/equine influenza (H3N8) viruses. J. Virol. Methods 20:1-12. - PubMed
-
- Crawford, P. C., E. J. Dubovi, W. L. Castleman, I. Stephenson, E. P. Gibbs, L. Chen, C. Smith, R. C. Hill, P. Ferro, J. Pompey, R. A. Bright, M. J. Medina, C. M. Johnson, C. W. Olsen, N. J. Cox, A. I. Klimov, J. M. Katz, and R. O. Donis. 2005. Transmission of equine influenza virus to dogs. Science 310:482-485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

