Ancient adaptive evolution of tetherin shaped the functions of Vpu and Nef in human immunodeficiency virus and primate lentiviruses
- PMID: 20444900
- PMCID: PMC2898239
- DOI: 10.1128/JVI.00468-10
Ancient adaptive evolution of tetherin shaped the functions of Vpu and Nef in human immunodeficiency virus and primate lentiviruses
Abstract
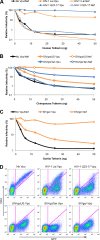
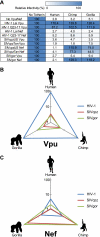
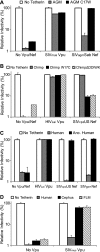
Tetherin/BST-2 is a host-encoded protein that restricts a wide diversity of viruses at the stage of virion release. However, viruses have evolved antagonists of Tetherin, including the Vpu and Nef proteins of primate lentiviruses. Like other host genes subject to viral antagonism, primate Tetherin genes have evolved under positive selection. We show here that viral antagonists acting at three independent sites of selection have driven the evolution of Tetherin, with the strongest selective pressure on the cytoplasmic tail domain. Human Tetherin is unique among the Tetherins of simian primates in that it has a 5-amino-acid deletion that results in the loss of the residue under the strongest positive selection. We show that this residue at amino acid 17 is the site of the functional interaction of Tetherin with Nef, since single amino acid substitutions at this single position can determine the susceptibility of Tetherin to Nef antagonism. While the simian immunodeficiency viruses SIVcpz and SIVgor are able to antagonize their hosts' Tetherin with Nef, human immunodeficiency virus type 1 (HIV-1) Vpu has evolved to counteract Tetherin in humans. We mapped the adaptations in the N-terminal transmembrane domain of Vpu that allow it to counteract human Tetherin. Our combined evolutionary and functional studies have allowed us to reconstruct the host-pathogen interactions that have shaped Tetherin and its lentivirus-encoded antagonists.
Figures



Similar articles
-
Counteraction of tetherin antiviral activity by two closely related SIVs differing by the presence of a Vpu gene.PLoS One. 2012;7(4):e35411. doi: 10.1371/journal.pone.0035411. Epub 2012 Apr 17. PLoS One. 2012. PMID: 22530020 Free PMC article.
-
Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2.PLoS Pathog. 2009 May;5(5):e1000429. doi: 10.1371/journal.ppat.1000429. Epub 2009 May 15. PLoS Pathog. 2009. PMID: 19436700 Free PMC article.
-
Vpu of a Simian Immunodeficiency Virus Isolated from Greater Spot-Nosed Monkey Antagonizes Human BST-2 via Two AxxxxxxxW Motifs.J Virol. 2020 Jan 6;94(2):e01669-19. doi: 10.1128/JVI.01669-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31666374 Free PMC article.
-
BST-2/tetherin: a new component of the innate immune response to enveloped viruses.Trends Microbiol. 2010 Sep;18(9):388-96. doi: 10.1016/j.tim.2010.06.010. Epub 2010 Aug 3. Trends Microbiol. 2010. PMID: 20688520 Free PMC article. Review.
-
Antiviral activity of the interferon-induced cellular protein BST-2/tetherin.AIDS Res Hum Retroviruses. 2009 Dec;25(12):1197-210. doi: 10.1089/aid.2009.0253. AIDS Res Hum Retroviruses. 2009. PMID: 19929170 Free PMC article. Review.
Cited by
-
Dual host-virus arms races shape an essential housekeeping protein.PLoS Biol. 2013;11(5):e1001571. doi: 10.1371/journal.pbio.1001571. Epub 2013 May 28. PLoS Biol. 2013. PMID: 23723737 Free PMC article.
-
Overlapping Patterns of Rapid Evolution in the Nucleic Acid Sensors cGAS and OAS1 Suggest a Common Mechanism of Pathogen Antagonism and Escape.PLoS Genet. 2015 May 5;11(5):e1005203. doi: 10.1371/journal.pgen.1005203. eCollection 2015 May. PLoS Genet. 2015. PMID: 25942676 Free PMC article.
-
Breaking Barriers to an AIDS Model with Macaque-Tropic HIV-1 Derivatives.Biology (Basel). 2012 May 12;1(2):134-64. doi: 10.3390/biology1020134. Biology (Basel). 2012. PMID: 23336082 Free PMC article.
-
Lack of adaptation to human tetherin in HIV-1 group O and P.Retrovirology. 2011 Sep 28;8:78. doi: 10.1186/1742-4690-8-78. Retrovirology. 2011. PMID: 21955466 Free PMC article.
-
Coevolutionary dynamics between tribe Cercopithecini tetherins and their lentiviruses.Sci Rep. 2015 Nov 4;5:16021. doi: 10.1038/srep16021. Sci Rep. 2015. PMID: 26531727 Free PMC article.
References
-
- Ariën, K., and B. Verhasselt. 2008. HIV Nef: role in pathogenesis and viral fitness. Curr. HIV Res. 6:200-208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

