Fusel alcohols regulate translation initiation by inhibiting eIF2B to reduce ternary complex in a mechanism that may involve altering the integrity and dynamics of the eIF2B body
- PMID: 20444979
- PMCID: PMC2893985
- DOI: 10.1091/mbc.e09-11-0962
Fusel alcohols regulate translation initiation by inhibiting eIF2B to reduce ternary complex in a mechanism that may involve altering the integrity and dynamics of the eIF2B body
Abstract
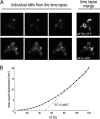
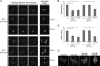
Recycling of eIF2-GDP to the GTP-bound form constitutes a core essential, regulated step in eukaryotic translation. This reaction is mediated by eIF2B, a heteropentameric factor with important links to human disease. eIF2 in the GTP-bound form binds to methionyl initiator tRNA to form a ternary complex, and the levels of this ternary complex can be a critical determinant of the rate of protein synthesis. Here we show that eIF2B serves as the target for translation inhibition by various fusel alcohols in yeast. Fusel alcohols are endpoint metabolites from amino acid catabolism, which signal nitrogen scarcity. We show that the inhibition of eIF2B leads to reduced ternary complex levels and that different eIF2B subunit mutants alter fusel alcohol sensitivity. A DNA tiling array strategy was developed that overcame difficulties in the identification of these mutants where the phenotypic distinctions were too subtle for classical complementation cloning. Fusel alcohols also lead to eIF2alpha dephosphorylation in a Sit4p-dependent manner. In yeast, eIF2B occupies a large cytoplasmic body where guanine nucleotide exchange on eIF2 can occur and be regulated. Fusel alcohols impact on both the movement and dynamics of this 2B body. Overall, these results confirm that the guanine nucleotide exchange factor, eIF2B, is targeted by fusel alcohols. Moreover, they highlight a potential connection between the movement or integrity of the 2B body and eIF2B regulation.
Figures


Similar articles
-
The beta/Gcd7 subunit of eukaryotic translation initiation factor 2B (eIF2B), a guanine nucleotide exchange factor, is crucial for binding eIF2 in vivo.Mol Cell Biol. 2010 Nov;30(21):5218-33. doi: 10.1128/MCB.00265-10. Epub 2010 Aug 30. Mol Cell Biol. 2010. PMID: 20805354 Free PMC article.
-
Identification of domains and residues within the epsilon subunit of eukaryotic translation initiation factor 2B (eIF2Bepsilon) required for guanine nucleotide exchange reveals a novel activation function promoted by eIF2B complex formation.Mol Cell Biol. 2000 Jun;20(11):3965-76. doi: 10.1128/MCB.20.11.3965-3976.2000. Mol Cell Biol. 2000. PMID: 10805739 Free PMC article.
-
eIF2B promotes eIF5 dissociation from eIF2*GDP to facilitate guanine nucleotide exchange for translation initiation.Genes Dev. 2013 Dec 15;27(24):2696-707. doi: 10.1101/gad.231514.113. Genes Dev. 2013. PMID: 24352424 Free PMC article.
-
Localization of the translational guanine nucleotide exchange factor eIF2B: a common theme for GEFs?Cell Cycle. 2006 Apr;5(7):678-80. doi: 10.4161/cc.5.7.2607. Epub 2006 Apr 1. Cell Cycle. 2006. PMID: 16582624 Review.
-
Clues to the mechanism of action of eIF2B, the guanine-nucleotide-exchange factor for translation initiation.Biochem Soc Trans. 2008 Aug;36(Pt 4):658-64. doi: 10.1042/BST0360658. Biochem Soc Trans. 2008. PMID: 18631136 Review.
Cited by
-
eIF2B localization and its regulation during the integrated stress response is cell-type specific.iScience. 2024 Aug 30;27(9):110851. doi: 10.1016/j.isci.2024.110851. eCollection 2024 Sep 20. iScience. 2024. PMID: 39310746 Free PMC article.
-
The yeast eukaryotic translation initiation factor 2B translation initiation complex interacts with the fatty acid synthesis enzyme YBR159W and endoplasmic reticulum membranes.Mol Cell Biol. 2013 Mar;33(5):1041-56. doi: 10.1128/MCB.00811-12. Epub 2012 Dec 21. Mol Cell Biol. 2013. PMID: 23263984 Free PMC article.
-
Alcohols inhibit translation to regulate morphogenesis in C. albicans.Fungal Genet Biol. 2015 Apr;77:50-60. doi: 10.1016/j.fgb.2015.03.008. Epub 2015 Apr 3. Fungal Genet Biol. 2015. PMID: 25843913 Free PMC article.
-
Glucose depletion inhibits translation initiation via eIF4A loss and subsequent 48S preinitiation complex accumulation, while the pentose phosphate pathway is coordinately up-regulated.Mol Biol Cell. 2011 Sep;22(18):3379-93. doi: 10.1091/mbc.E11-02-0153. Epub 2011 Jul 27. Mol Biol Cell. 2011. PMID: 21795399 Free PMC article.
-
Dynamic changes in eIF4F-mRNA interactions revealed by global analyses of environmental stress responses.Genome Biol. 2017 Oct 27;18(1):201. doi: 10.1186/s13059-017-1338-4. Genome Biol. 2017. PMID: 29078784 Free PMC article.
References
-
- Burns N., Grimwade B., Rossmacdonald P. B., Choi E. Y., Finberg K., Roeder G. S., Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption Saccharomyces cerevisiae. Gene Dev. 1994;8:1087–1105. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

