Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae
- PMID: 20444982
- PMCID: PMC2893990
- DOI: 10.1091/mbc.e09-04-0345
Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae
Abstract
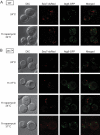
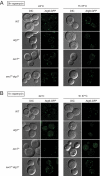
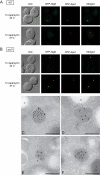
The delivery of proteins and organelles to the vacuole by autophagy involves membrane rearrangements that result in the formation of large vesicles called autophagosomes. The mechanism underlying autophagosome biogenesis and the origin of the membranes composing these vesicles remains largely unclear. We have investigated the role of the Golgi complex in autophagy and have determined that in yeast, activation of ADP-ribosylation factor (Arf)1 and Arf2 GTPases by Sec7, Gea1, and Gea2 is essential for this catabolic process. The two main events catalyzed by these components, the biogenesis of COPI- and clathrin-coated vesicles, do not play a critical role in autophagy. Analysis of the sec7 strain under starvation conditions revealed that the autophagy machinery is correctly assembled and the precursor membrane cisterna of autophagosomes, the phagophore, is normally formed. However, the expansion of the phagophore into an autophagosome is severely impaired. Our data show that the Golgi complex plays a crucial role in supplying the lipid bilayers necessary for the biogenesis of double-membrane vesicles possibly through a new class of transport carriers or a new mechanism.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

