Colloquium paper: genome-wide patterns of population structure and admixture among Hispanic/Latino populations
- PMID: 20445096
- PMCID: PMC3024022
- DOI: 10.1073/pnas.0914618107
Colloquium paper: genome-wide patterns of population structure and admixture among Hispanic/Latino populations
Abstract
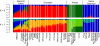
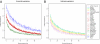
Hispanic/Latino populations possess a complex genetic structure that reflects recent admixture among and potentially ancient substructure within Native American, European, and West African source populations. Here, we quantify genome-wide patterns of SNP and haplotype variation among 100 individuals with ancestry from Ecuador, Colombia, Puerto Rico, and the Dominican Republic genotyped on the Illumina 610-Quad arrays and 112 Mexicans genotyped on Affymetrix 500K platform. Intersecting these data with previously collected high-density SNP data from 4,305 individuals, we use principal component analysis and clustering methods FRAPPE and STRUCTURE to investigate genome-wide patterns of African, European, and Native American population structure within and among Hispanic/Latino populations. Comparing autosomal, X and Y chromosome, and mtDNA variation, we find evidence of a significant sex bias in admixture proportions consistent with disproportionate contribution of European male and Native American female ancestry to present-day populations. We also find that patterns of linkage-disequilibria in admixed Hispanic/Latino populations are largely affected by the admixture dynamics of the populations, with faster decay of LD in populations of higher African ancestry. Finally, using the locus-specific ancestry inference method LAMP, we reconstruct fine-scale chromosomal patterns of admixture. We document moderate power to differentiate among potential subcontinental source populations within the Native American, European, and African segments of the admixed Hispanic/Latino genomes. Our results suggest future genome-wide association scans in Hispanic/Latino populations may require correction for local genomic ancestry at a subcontinental scale when associating differences in the genome with disease risk, progression, and drug efficacy, as well as for admixture mapping.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



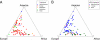
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

