Conformational plasticity and structure/function relationships in cytochromes P450
- PMID: 20446763
- PMCID: PMC2959183
- DOI: 10.1089/ars.2010.3109
Conformational plasticity and structure/function relationships in cytochromes P450
Abstract
The cytochrome P450s are a superfamily of enzymes that are found in all kingdoms of living organisms, and typically catalyze the oxidative addition of atomic oxygen to an unactivated C-C or C-H bond. Over 8000 nonredundant sequences of putative and confirmed P450 enzymes have been identified, but three-dimensional structures have been determined for only a small fraction of these. While all P450 enzymes for which structures have been determined share a common global fold, the flexibility and modularity of structure around the active site account for the ability of P450 enzymes to accommodate a vast number of structurally dissimilar substrates and support a wide range of selective oxidations. In this review, known P450 structures are compared, and some structural criteria for prediction of substrate selectivity and reaction type are suggested. The importance of dynamic processes such as redox-dependent and effector-induced conformational changes in determining catalytic competence and regio- and stereoselectivity is discussed, and noncrystallographic methods for characterizing P450 structures and dynamics, in particular, mass spectrometry and nuclear magnetic resonance spectroscopy are reviewed.
Figures
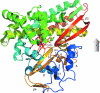

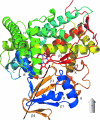

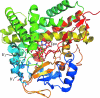

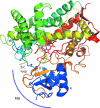











Similar articles
-
What Your Crystal Structure Will Not Tell You about Enzyme Function.Acc Chem Res. 2019 May 21;52(5):1409-1418. doi: 10.1021/acs.accounts.9b00066. Epub 2019 Apr 29. Acc Chem Res. 2019. PMID: 31034199 Free PMC article.
-
Detection of substrate-dependent conformational changes in the P450 fold by nuclear magnetic resonance.Sci Rep. 2016 Feb 25;6:22035. doi: 10.1038/srep22035. Sci Rep. 2016. PMID: 26911901 Free PMC article.
-
Future perception in P450 research.J Inorg Biochem. 2018 Sep;186:264-266. doi: 10.1016/j.jinorgbio.2018.06.002. Epub 2018 Jun 7. J Inorg Biochem. 2018. PMID: 29990750
-
Use of chemical auxiliaries to control p450 enzymes for predictable oxidations at unactivated C-h bonds of substrates.Adv Exp Med Biol. 2015;851:209-28. doi: 10.1007/978-3-319-16009-2_8. Adv Exp Med Biol. 2015. PMID: 26002737 Free PMC article. Review.
-
P450BM-3; a tale of two domains--or is it three?Steroids. 1997 Jan;62(1):117-23. doi: 10.1016/s0039-128x(96)00169-9. Steroids. 1997. PMID: 9029725 Review.
Cited by
-
Cafestol to Tricalysiolide B and Oxidized Analogues: Biosynthetic and Derivatization Studies Using Non-heme Iron Catalyst Fe(PDP).Synlett. 2012 Dec 1;23(19):2768-2772. doi: 10.1055/s-0032-1317708. Synlett. 2012. PMID: 23585710 Free PMC article.
-
Identification of Genes Encoding Enzymes Catalyzing the Early Steps of Carrot Polyacetylene Biosynthesis.Plant Physiol. 2018 Dec;178(4):1507-1521. doi: 10.1104/pp.18.01195. Epub 2018 Oct 17. Plant Physiol. 2018. PMID: 30333150 Free PMC article.
-
How can SHAP values help to shape metabolic stability of chemical compounds?J Cheminform. 2021 Sep 27;13(1):74. doi: 10.1186/s13321-021-00542-y. J Cheminform. 2021. PMID: 34579792 Free PMC article.
-
Conformational Mobility in Cytochrome P450 3A4 Explored by Pressure-Perturbation EPR Spectroscopy.Biophys J. 2016 Apr 12;110(7):1485-1498. doi: 10.1016/j.bpj.2016.02.026. Biophys J. 2016. PMID: 27074675 Free PMC article.
-
Point Mutations at a Key Site Alter the Cytochrome P450 OleP Structural Dynamics.Biomolecules. 2021 Dec 31;12(1):55. doi: 10.3390/biom12010055. Biomolecules. 2021. PMID: 35053203 Free PMC article.
References
-
- Andersen JF. Tatsuta K. Gunji H. Ishiyama T. Hutchinson CR. Substrate-apecificity of 6-Deoxyerythronolide-B hydroxylase, a bacterial cytochrome P450 of erythromycin-a biosynthesis. Biochemistry. 1993;32:1905–1913. - PubMed
-
- Arimoto R. Computational models for predicting interactions with cytochrome P450 enzyme. Current Topics Med Chem. 2006;6:1609–1618. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

