Regulation of PKR by HCV IRES RNA: importance of domain II and NS5A
- PMID: 20447405
- PMCID: PMC2902579
- DOI: 10.1016/j.jmb.2010.04.059
Regulation of PKR by HCV IRES RNA: importance of domain II and NS5A
Abstract
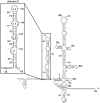
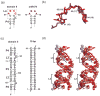
Protein kinase R (PKR) is an essential component of the innate immune response. In the presence of double-stranded RNA (dsRNA), PKR is autophosphorylated, which enables it to phosphorylate its substrate, eukaryotic initiation factor 2alpha, leading to translation cessation. Typical activators of PKR are long dsRNAs produced during viral infection, although certain other RNAs can also activate. A recent study indicated that full-length internal ribosome entry site (IRES), present in the 5'-untranslated region of hepatitis C virus (HCV) RNA, inhibits PKR, while another showed that it activates. We show here that both activation and inhibition by full-length IRES are possible. The HCV IRES has a complex secondary structure comprising four domains. While it has been demonstrated that domains III-IV activate PKR, we report here that domain II of the IRES also potently activates. Structure mapping and mutational analysis of domain II indicate that while the double-stranded regions of the RNA are important for activation, loop regions contribute as well. Structural comparison reveals that domain II has multiple, non-Watson-Crick features that mimic A-form dsRNA. The canonical and noncanonical features of domain II cumulate to a total of approximately 33 unbranched base pairs, the minimum length of dsRNA required for PKR activation. These results provide further insight into the structural basis of PKR activation by a diverse array of RNA structural motifs that deviate from the long helical stretches found in traditional PKR activators. Activation of PKR by domain II of the HCV IRES has implications for the innate immune response when the other domains of the IRES may be inaccessible. We also study the ability of the HCV nonstructural protein 5A (NS5A) to bind various domains of the IRES and alter activation. A model is presented for how domain II of the IRES and NS5A operate to control host and viral translation during HCV infection.
2010 Elsevier Ltd. All rights reserved.
Figures








References
-
- Balachandran S, Roberts PC, Brown LE, Truong H, Pattnaik AK, Archer DR, Barber GN. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity. 2000;13:129–141. - PubMed
-
- Williams BR. PKR; a sentinel kinase for cellular stress. Oncogene. 1999;18:6112–6120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

