Advances in RNA structure analysis by chemical probing
- PMID: 20447823
- PMCID: PMC2916962
- DOI: 10.1016/j.sbi.2010.04.001
Advances in RNA structure analysis by chemical probing
Abstract
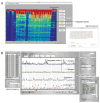
RNA is arguably the most versatile biological macromolecule because of its ability both to encode and to manipulate genetic information. The diverse roles of RNA depend on its ability to fold back on itself to form biologically functional structures that bind small molecule and large protein ligands, to change conformation, and to affect the cellular regulatory state. These features of RNA biology can be structurally interrogated using chemical mapping experiments. The usefulness and applications of RNA chemical probing technologies have expanded dramatically over the past five years because of several critical advances. These innovations include new sequence-independent RNA chemistries, algorithmic tools for high-throughput analysis of complex data sets composed of thousands of measurements, new approaches for interpreting chemical probing data for both secondary and tertiary structure prediction, facile methods for following time-dependent processes, and the willingness of individual research groups to tackle increasingly bold problems in RNA structural biology.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures




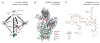
References
-
- Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. - PubMed
-
- Lavery R, Pullman A. A New Theoretical Index of Biochemical Reactivity Combining Steric and Electrostatic Factors. Biophys Chem. 1984;19:171–181. - PubMed
-
- Tullius TD, Greenbaum JA. Mapping nucleic acid structure by hydroxyl radical cleavage. Curr Opin Struct Biol. 2005;9:127–134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

