The mechanism and control of DNA transfer by the conjugative relaxase of resistance plasmid pCU1
- PMID: 20448025
- PMCID: PMC2943615
- DOI: 10.1093/nar/gkq303
The mechanism and control of DNA transfer by the conjugative relaxase of resistance plasmid pCU1
Abstract
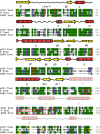
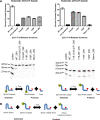
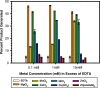
Bacteria expand their genetic diversity, spread antibiotic resistance genes, and obtain virulence factors through the highly coordinated process of conjugative plasmid transfer (CPT). A plasmid-encoded relaxase enzyme initiates and terminates CPT by nicking and religating the transferred plasmid in a sequence-specific manner. We solved the 2.3 A crystal structure of the relaxase responsible for the spread of the resistance plasmid pCU1 and determined its DNA binding and nicking capabilities. The overall fold of the pCU1 relaxase is similar to that of the F plasmid and plasmid R388 relaxases. However, in the pCU1 structure, the conserved tyrosine residues (Y18,19,26,27) that are required for DNA nicking and religation were displaced up to 14 A out of the relaxase active site, revealing a high degree of mobility in this region of the enzyme. In spite of this flexibility, the tyrosines still cleaved the nic site of the plasmid's origin of transfer, and did so in a sequence-specific, metal-dependent manner. Unexpectedly, the pCU1 relaxase lacked the sequence-specific DNA binding previously reported for the homologous F and R388 relaxase enzymes, despite its high sequence and structural similarity with both proteins. In summary, our work outlines novel structural and functional aspects of the relaxase-mediated conjugative transfer of plasmid pCU1.
Figures

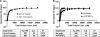
Similar articles
-
Tyrosine partners coordinate DNA nicking by the Salmonella typhimurium plasmid pCU1 relaxase enzyme.FEBS Lett. 2011 Apr 20;585(8):1216-22. doi: 10.1016/j.febslet.2011.03.043. Epub 2011 Mar 23. FEBS Lett. 2011. PMID: 21439279 Free PMC article.
-
Catalytic domain of plasmid pAD1 relaxase TraX defines a group of relaxases related to restriction endonucleases.Proc Natl Acad Sci U S A. 2013 Aug 13;110(33):13606-11. doi: 10.1073/pnas.1310037110. Epub 2013 Jul 31. Proc Natl Acad Sci U S A. 2013. PMID: 23904483 Free PMC article.
-
A high security double lock and key mechanism in HUH relaxases controls oriT-processing for plasmid conjugation.Nucleic Acids Res. 2014;42(16):10632-43. doi: 10.1093/nar/gku741. Epub 2014 Aug 14. Nucleic Acids Res. 2014. PMID: 25123661 Free PMC article.
-
Nicking by transesterification: the reaction catalysed by a relaxase.Mol Microbiol. 1997 Sep;25(6):1011-22. doi: 10.1046/j.1365-2958.1997.5241885.x. Mol Microbiol. 1997. PMID: 9350859 Review.
-
The diversity of conjugative relaxases and its application in plasmid classification.FEMS Microbiol Rev. 2009 May;33(3):657-87. doi: 10.1111/j.1574-6976.2009.00168.x. FEMS Microbiol Rev. 2009. PMID: 19396961 Review.
Cited by
-
Investigating the impact of bisphosphonates and structurally related compounds on bacteria containing conjugative plasmids.Biochem Biophys Res Commun. 2012 Aug 10;424(4):697-703. doi: 10.1016/j.bbrc.2012.07.012. Epub 2012 Jul 13. Biochem Biophys Res Commun. 2012. PMID: 22796221 Free PMC article.
-
Orthogonal Protein Assembly on DNA Nanostructures Using Relaxases.Angew Chem Int Ed Engl. 2016 Mar 18;55(13):4348-52. doi: 10.1002/anie.201510313. Epub 2016 Feb 24. Angew Chem Int Ed Engl. 2016. PMID: 26915475 Free PMC article.
-
Structural and functional characterization of TraI from pKM101 reveals basis for DNA processing.Life Sci Alliance. 2023 Jan 20;6(4):e202201775. doi: 10.26508/lsa.202201775. Print 2023 Apr. Life Sci Alliance. 2023. PMID: 36669792 Free PMC article.
-
Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.Proc Natl Acad Sci U S A. 2017 Aug 8;114(32):E6526-E6535. doi: 10.1073/pnas.1702971114. Epub 2017 Jul 24. Proc Natl Acad Sci U S A. 2017. PMID: 28739894 Free PMC article.
-
DNA processing by the MOBH family relaxase TraI encoded within the gonococcal genetic island.Nucleic Acids Res. 2019 Sep 5;47(15):8136-8153. doi: 10.1093/nar/gkz577. Nucleic Acids Res. 2019. PMID: 31276596 Free PMC article.
References
-
- Siegel JD, Rhinehart E, Jackson M, Chiarello L. The Centers for Disease Control and Prevention. Healthcare Infection Control Practices Advisory Committee; 2006. Management of Multidrug-Resistant Organisms in Healthcare Settings. - PubMed
-
- Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 2007;3:541–548. - PubMed
-
- Fernandez-Lopez R, Machon C, Longshaw CM, Martin S, Molin S, Zechner EL, Espinosa M, Lanka E, de la Cruz F. Unsaturated fatty acids are inhibitors of bacterial conjugation. Microbiology. 2005;151:3517–3626. - PubMed
-
- Garcillan-Barcia MP, Jurado P, Gonzalez-Perez B, Moncalian G, Fernandez LA, de la Cruz F. Conjugative transfer can be inhibited by blocking relaxase activity within recipient cells with intrabodies. Mol. Microbiol. 2007;63:404–416. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

