Electrostatic suppression allows tyrosine site-specific recombination in the absence of a conserved catalytic arginine
- PMID: 20448041
- PMCID: PMC2906291
- DOI: 10.1074/jbc.M110.112292
Electrostatic suppression allows tyrosine site-specific recombination in the absence of a conserved catalytic arginine
Abstract
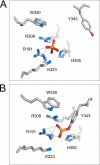
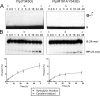
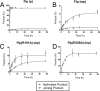
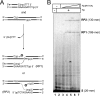
The active site of the tyrosine family site-specific recombinase Flp contains a conserved catalytic pentad that includes two arginine residues, Arg-191 and Arg-308. Both arginines are essential for the transesterification steps of strand cleavage and strand joining in DNA substrates containing a phosphate group at the scissile position. During strand cleavage, the active site tyrosine supplies the nucleophile to form a covalent 3'-phosphotyrosyl intermediate. The 5'-hydroxyl group produced by cleavage provides the nucleophile to re-form a 3'-5' phosphodiester bond in a recombinant DNA strand. In previous work we showed that substitution of the scissile phosphate (P) by the charge neutral methylphosphonate (MeP) makes Arg-308 dispensable during the catalytic activation of the MeP diester bond. However, in the Flp(R308A) reaction, water out-competes the tyrosine nucleophile (Tyr-343) to cause direct hydrolysis of the MeP diester bond. We now report that for MeP activation Arg-191 is also not required. In contrast to Flp(R308A), Flp(R191A) primarily mediates normal cleavage by Tyr-343 but also exhibits a weaker direct hydrolytic activity. The cleaved MeP-tyrosyl intermediate formed by Flp(R191A) can be targeted for nucleophilic attack by a 5'-hydroxyl or water and channeled toward strand joining or hydrolysis, respectively. In collaboration with wild type Flp, Flp(R191A) promotes strand exchange between MeP- and P-DNA partners. Loss of a catalytically crucial positively charged side chain can thus be suppressed by a compensatory modification in the DNA substrate that neutralizes the negative charge on the scissile phosphate.
Figures



References
-
- Perry K., Hwang Y., Bushman F. D., Van Duyne G. D. (2006) Mol. Cell 23, 343–354 - PubMed
-
- Grindley N. D., Whiteson K. L., Rice P. A. (2006) Annu. Rev. Biochem. 75, 567–605 - PubMed
-
- Chen Y., Rice P. A. (2003) Annu. Rev. Biophys. Biomol. Struct. 32, 135–159 - PubMed
-
- Grainge I., Jayaram M. (1999) Mol. Microbiol. 33, 449–456 - PubMed
-
- Guo F., Gopaul D. N., van Duyne G. D. (1997) Nature 389, 40–46 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

