Peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC-1alpha) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis
- PMID: 20448046
- PMCID: PMC2898414
- DOI: 10.1074/jbc.M109.070169
Peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC-1alpha) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis
Abstract
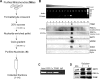
The transcriptional co-activator PGC-1alpha and the NAD(+)-dependent deacetylase SIRT1 are considered important inducers of mitochondrial biogenesis because in the nucleus they regulate transcription of nucleus-encoded mitochondrial genes. We demonstrate that PGC-1alpha and SIRT1 are also present inside mitochondria and are in close proximity to mtDNA. They interact with mitochondrial transcription factor A (TFAM) as assessed by confocal microscopy analysis and by blue native-PAGE. Nucleoid purification allowed us to identify SIRT1 and PGC-1alpha as proteins associated with native and cross-linked nucleoids, respectively. After mtDNA immunoprecipitation analysis, carried out on mitochondrial extracts, we found that PGC-1alpha is present on the same D-loop region recognized by TFAM. Finally, by oligonucleotide pulldown assay, we found PGC-1alpha and SIRT1 associated with the TFAM consensus sequence (human mitochondrial transcription factor-binding site H). The results obtained suggest that in mitochondria PGC-1alpha and SIRT1 may function as their nuclear counterparts and represent the genuine factors mediating the cross-talk between nuclear and mitochondrial genome. Finally, this work adds new knowledge on the function of SIRT1 and PGC-1alpha and highlights the direct involvement of such proteins in regulation of mitochondrial biogenesis.
Figures
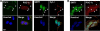




References
-
- Hock M. B., Kralli A. (2009) Annu. Rev. Physiol. 71, 177–203 - PubMed
-
- Chow L. S., Greenlund L. J., Asmann Y. W., Short K. R., McCrady S. K., Levine J. A., Nair K. S. (2007) J. Appl. Physiol. 102, 1078–1089 - PubMed
-
- Scarpulla R. C. (2008) Physiol. Rev. 88, 611–638 - PubMed
-
- Guarente L. (2007) Cold Spring Harbor Symp. Quant. Biol. 72, 483–488 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

