The genetic basis of kidney cancer: a metabolic disease
- PMID: 20448661
- PMCID: PMC2929006
- DOI: 10.1038/nrurol.2010.47
The genetic basis of kidney cancer: a metabolic disease
Abstract
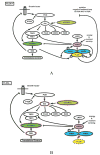
Kidney cancer is not a single disease but comprises a number of different types of cancer that occur in the kidney, each caused by a different gene with a different histology and clinical course that responds differently to therapy. Each of the seven known kidney cancer genes, VHL, MET, FLCN, TSC1, TSC2, FH and SDH, is involved in pathways that respond to metabolic stress or nutrient stimulation. The VHL protein is a component of the oxygen and iron sensing pathway that regulates hypoxia-inducible factor (HIF) levels in the cell. HGF-MET signaling affects the LKB1-AMPK energy sensing cascade. The FLCN-FNIP1-FNIP2 complex binds AMPK and, therefore, might interact with the cellular energy and nutrient sensing pathways AMPK-TSC1/2-mTOR and PI3K-Akt-mTOR. TSC1-TSC2 is downstream of AMPK and negatively regulates mTOR in response to cellular energy deficit. FH and SDH have a central role in the mitochondrial tricarboxylic acid cycle, which is coupled to energy production through oxidative phosphorylation. Mutations in each of these kidney cancer genes result in dysregulation of metabolic pathways involved in oxygen, iron, energy or nutrient sensing, suggesting that kidney cancer is a disease of cell metabolism. Targeting the fundamental metabolic abnormalities in kidney cancer provides a unique opportunity for the development of more-effective forms of therapy for this disease.
Conflict of interest statement
Competing interests
The authors have no competing interests
Figures






References
-
- Linehan WM, Walther MM, Zbar B. The genetic basis of cancer of the kidney. J Urol. 2003 Dec;170(6 Pt 1):2163–72. - PubMed
-
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004 Aug;10(8):789–99. - PubMed
-
- Thompson CB. Attacking cancer at its root. Cell. 2009 Sep 18;138(6):1051–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

