Learning patient-specific predictive models from clinical data
- PMID: 20450985
- PMCID: PMC2933959
- DOI: 10.1016/j.jbi.2010.04.009
Learning patient-specific predictive models from clinical data
Abstract
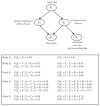
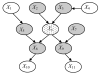
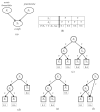
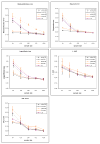
We introduce an algorithm for learning patient-specific models from clinical data to predict outcomes. Patient-specific models are influenced by the particular history, symptoms, laboratory results, and other features of the patient case at hand, in contrast to the commonly used population-wide models that are constructed to perform well on average on all future cases. The patient-specific algorithm uses Markov blanket (MB) models, carries out Bayesian model averaging over a set of models to predict the outcome for the patient case at hand, and employs a patient-specific heuristic to locate a set of suitable models to average over. We evaluate the utility of using a local structure representation for the conditional probability distributions in the MB models that captures additional independence relations among the variables compared to the typically used representation that captures only the global structure among the variables. In addition, we compare the performance of Bayesian model averaging to that of model selection. The patient-specific algorithm and its variants were evaluated on two clinical datasets for two outcomes. Our results provide support that the performance of an algorithm for learning patient-specific models can be improved by using a local structure representation for MB models and by performing Bayesian model averaging.
Figures





References
-
- van Bemmel JH, Musen MA. Handbook of Medical Informatics. 1. New York: Springer-Verlag; 1997.
-
- Abu-Hanna A, Lucas PJ. Prognostic models in medicine. AI and statistical approaches. Methods of Information in Medicine. 2001 Mar;40(1):1–5. - PubMed
-
- Cooper GF, Aliferis CF, Ambrosino R, Aronis J, Buchanan BG, Caruana R, et al. An evaluation of machine-learning methods for predicting pneumonia mortality. Artificial Intelligence. 1997 Feb;9(2):107–38. - PubMed
-
- Hoeting JA, Madigan D, Raftery AE, Volinsky CT. Bayesian model averaging: A tutorial. Statistical Science. 1999 Nov;14(4):382–401.
-
- Visweswaran S. PhD dissertation. Pittsburgh: University of Pittsburgh; 2007. Learning patient-specific models from clinical data. [updated 2007; cited]; Available from: http://etd.library.pitt.edu/ETD/available/etd-11292007-232406/unrestrict....
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

