Peroxisomes are signaling platforms for antiviral innate immunity
- PMID: 20451243
- PMCID: PMC3670185
- DOI: 10.1016/j.cell.2010.04.018
Peroxisomes are signaling platforms for antiviral innate immunity
Abstract
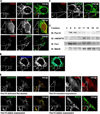
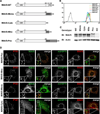
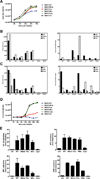
Peroxisomes have long been established to play a central role in regulating various metabolic activities in mammalian cells. These organelles act in concert with mitochondria to control the metabolism of lipids and reactive oxygen species. However, while mitochondria have emerged as an important site of antiviral signal transduction, a role for peroxisomes in immune defense is unknown. Here, we report that the RIG-I-like receptor (RLR) adaptor protein MAVS is located on peroxisomes and mitochondria. We find that peroxisomal and mitochondrial MAVS act sequentially to create an antiviral cellular state. Upon viral infection, peroxisomal MAVS induces the rapid interferon-independent expression of defense factors that provide short-term protection, whereas mitochondrial MAVS activates an interferon-dependent signaling pathway with delayed kinetics, which amplifies and stabilizes the antiviral response. The interferon regulatory factor IRF1 plays a crucial role in regulating MAVS-dependent signaling from peroxisomes. These results establish that peroxisomes are an important site of antiviral signal transduction.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Viral defense: it takes two MAVS to Tango.Cell. 2010 May 14;141(4):570-2. doi: 10.1016/j.cell.2010.04.043. Cell. 2010. PMID: 20478250
-
Antiviral immunity: Speed and endurance required.Nat Rev Immunol. 2010 Jul;10(7):465. doi: 10.1038/nri2809. Nat Rev Immunol. 2010. PMID: 20586123 No abstract available.
References
-
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. - PubMed
-
- Balachandran S, Venkataraman T, Fisher PB, Barber GN. Fas-associated death domain-containing protein-mediated antiviral innate immune signaling involves the regulation of Irf7. J Immunol. 2007;178:2429–2439. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

