The oncogenic phosphatase WIP1 negatively regulates nucleotide excision repair
- PMID: 20451471
- PMCID: PMC2893264
- DOI: 10.1016/j.dnarep.2010.04.005
The oncogenic phosphatase WIP1 negatively regulates nucleotide excision repair
Abstract
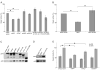
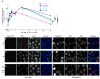
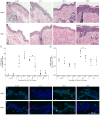
Nucleotide excision repair (NER) is the only mechanism in humans to repair UV-induced DNA lesions such as pyrimidine (6-4) pyrimidone photoproducts and cyclobutane pyrimidine dimers (CPDs). In response to UV damage, the ataxia telangiectasia mutated and Rad3-related (ATR) kinase phosphorylates and activates several downstream effector proteins, such as p53 and XPA, to arrest cell cycle progression, stimulate DNA repair, or initiate apoptosis. However, following the completion of DNA repair, there must be active mechanisms that restore the cell to a prestressed homeostatic state. An important part of this recovery must include a process to reduce p53 and NER activity as well as to remove repair protein complexes from the DNA damage sites. Since activation of the damage response occurs in part through phosphorylation, phosphatases are obvious candidates as homeostatic regulators of the DNA damage and repair responses. Therefore, we investigated whether the serine/threonine wild-type p53-induced phosphatase 1 (WIP1/PPM1D) might regulate NER. WIP1 overexpression inhibits the kinetics of NER and CPD repair, whereas WIP1 depletion enhances NER kinetics and CPD repair. This NER suppression is dependent on WIP1 phosphatase activity, as phosphatase-dead WIP1 mutants failed to inhibit NER. Moreover, WIP1 suppresses the kinetics of UV-induced damage repair largely through effects on NER, as XPD-deficient cells are not further suppressed in repairing UV damage by overexpressed WIP1. Wip1 null mice quickly repair their CPD and undergo less UV-induced apoptosis than their wild-type counterparts. In vitro phosphatase assays identify XPA and XPC as two potential WIP1 targets in the NER pathway. Thus WIP1 may suppress NER kinetics by dephosphorylating and inactivating XPA and XPC and other NER proteins and regulators after UV-induced DNA damage is repaired.
2010 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
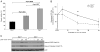
Similar articles
-
Checkpoint kinase ATR promotes nucleotide excision repair of UV-induced DNA damage via physical interaction with xeroderma pigmentosum group A.J Biol Chem. 2009 Sep 4;284(36):24213-22. doi: 10.1074/jbc.M109.000745. Epub 2009 Jul 8. J Biol Chem. 2009. PMID: 19586908 Free PMC article.
-
Phosphorylation of nucleotide excision repair factor xeroderma pigmentosum group A by ataxia telangiectasia mutated and Rad3-related-dependent checkpoint pathway promotes cell survival in response to UV irradiation.Cancer Res. 2006 Mar 15;66(6):2997-3005. doi: 10.1158/0008-5472.CAN-05-3403. Cancer Res. 2006. PMID: 16540648 Free PMC article.
-
UV-induced histone H2AX phosphorylation and DNA damage related proteins accumulate and persist in nucleotide excision repair-deficient XP-B cells.DNA Repair (Amst). 2011 Jan 2;10(1):5-15. doi: 10.1016/j.dnarep.2010.09.004. Epub 2010 Oct 13. DNA Repair (Amst). 2011. PMID: 20947453 Free PMC article.
-
Xeroderma Pigmentosa Group A (XPA), Nucleotide Excision Repair and Regulation by ATR in Response to Ultraviolet Irradiation.Adv Exp Med Biol. 2017;996:41-54. doi: 10.1007/978-3-319-56017-5_4. Adv Exp Med Biol. 2017. PMID: 29124689 Free PMC article. Review.
-
The type 2C phosphatase Wip1: an oncogenic regulator of tumor suppressor and DNA damage response pathways.Cancer Metastasis Rev. 2008 Jun;27(2):123-35. doi: 10.1007/s10555-008-9127-x. Cancer Metastasis Rev. 2008. PMID: 18265945 Free PMC article. Review.
Cited by
-
Activation of WIP1 phosphatase by HTLV-1 Tax mitigates the cellular response to DNA damage.PLoS One. 2013;8(2):e55989. doi: 10.1371/journal.pone.0055989. Epub 2013 Feb 6. PLoS One. 2013. PMID: 23405243 Free PMC article.
-
Nucleotide Excision Repair Pathway Activity Is Inhibited by Airborne Particulate Matter (PM10) through XPA Deregulation in Lung Epithelial Cells.Int J Mol Sci. 2022 Feb 17;23(4):2224. doi: 10.3390/ijms23042224. Int J Mol Sci. 2022. PMID: 35216341 Free PMC article.
-
SOD1 is a synthetic lethal target in PPM1D-mutant leukemia cells.bioRxiv [Preprint]. 2024 Jan 17:2023.08.31.555634. doi: 10.1101/2023.08.31.555634. bioRxiv. 2024. Update in: Elife. 2024 Jun 18;12:RP91611. doi: 10.7554/eLife.91611. PMID: 37693622 Free PMC article. Updated. Preprint.
-
Molecular determinants of ovarian cancer chemoresistance: new insights into an old conundrum.Ann N Y Acad Sci. 2012 Oct;1271(1):58-67. doi: 10.1111/j.1749-6632.2012.06734.x. Ann N Y Acad Sci. 2012. PMID: 23050965 Free PMC article. Review.
-
Sirtuin 1-mediated deacetylation of XPA DNA repair protein enhances its interaction with ATR protein and promotes cAMP-induced DNA repair of UV damage.J Biol Chem. 2018 Dec 7;293(49):19025-19037. doi: 10.1074/jbc.RA118.003940. Epub 2018 Oct 16. J Biol Chem. 2018. Retraction in: J Biol Chem. 2021 Jan-Jun;296:100185. doi: 10.1016/j.jbc.2020.100185. PMID: 30327428 Free PMC article. Retracted.
References
-
- Gillet LCJ, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chemical Reviews. 2006;106:253–276. - PubMed
-
- Friedberg EC. How nucleotide excision repair protects against cancer. Nat Rev Cancer. 2001;1:22–33. - PubMed
-
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. - PubMed
-
- Lazzaro F, Giannattasio M, Puddu F, Granata M, Pellicioli A, Plevani P, Muzi-Falconi M. Checkpoint mechanisms at the intersection between DNA damage and repair. DNA Repair (Amst) 2009;8:1055–1067. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

