The macroscopic rate of nucleic acid translocation by hepatitis C virus helicase NS3h is dependent on both sugar and base moieties
- PMID: 20451531
- PMCID: PMC2902667
- DOI: 10.1016/j.jmb.2010.04.065
The macroscopic rate of nucleic acid translocation by hepatitis C virus helicase NS3h is dependent on both sugar and base moieties
Abstract
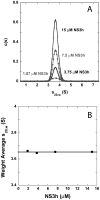
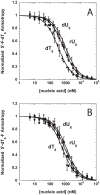
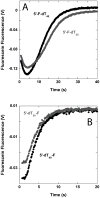
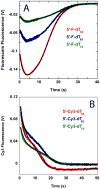
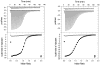
The nonstructural protein 3 helicase (NS3h) of hepatitis C virus is a 3'-to-5' superfamily 2 RNA and DNA helicase that is essential for the replication of hepatitis C virus. We have examined the kinetic mechanism of the translocation of NS3h along single-stranded nucleic acid with bases uridylate (rU), deoxyuridylate (dU), and deoxythymidylate (dT), and have found that the macroscopic rate of translocation is dependent on both the base moiety and the sugar moiety of the nucleic acid, with approximate macroscopic translocation rates of 3 nt s(-1) (oligo(dT)), 35 nt s(-1) (oligo(dU)), and 42 nt s(-1) (oligo(rU)), respectively. We found a strong correlation between the macroscopic translocation rates and the binding affinity of the translocating NS3h protein for the respective substrates such that weaker affinity corresponded to faster translocation. The values of K(0.5) for NS3h translocation at a saturating ATP concentration are as follows: 3.3+/-0.4 microM nucleotide (poly(dT)), 27+/-2 microM nucleotide (poly(dU)), and 36+/-2 microM nucleotide (poly(rU)). Furthermore, results of the isothermal titration of NS3h with these oligonucleotides suggest that differences in TDeltaS(0) are the principal source of differences in the affinity of NS3h binding to these substrates. Interestingly, despite the differences in macroscopic translocation rates and binding affinities, the ATP coupling stoichiometries for NS3h translocation were identical for all three substrates (approximately 0.5 ATP molecule consumed per nucleotide translocated). This similar periodicity of ATP consumption implies a similar mechanism for NS3h translocation along RNA and DNA substrates.
2010 Elsevier Ltd. All rights reserved.
Figures








References
-
- LOHMAN TM, BJORNSON KP. MECHANISMS OF HELICASE-CATALYZED DNA UNWINDING. ANNU REV BIOCHEM. 1996;65:169–214. - PubMed
-
- LOHMAN TM, TOMKO EJ, WU CG. NON-HEXAMERIC DNA HELICASES AND TRANSLOCASES: MECHANISMS AND REGULATION. NAT REV MOL CELL BIOL. 2008;9:391–401. - PubMed
-
- PATEL SS, DONMEZ I. MECHANISMS OF HELICASES. J BIOL CHEM. 2006;281:18265–8. - PubMed
-
- PYLE AM. TRANSLOCATION AND UNWINDING MECHANISMS OF RNA AND DNA HELICASES. ANNU REV BIOPHYS. 2008;37:317–36. - PubMed
-
- SINGLETON MR, DILLINGHAM MS, WIGLEY DB. STRUCTURE AND MECHANISM OF HELICASES AND NUCLEIC ACID TRANSLOCASES. ANNU REV BIOCHEM. 2007;76:23–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

