Distinct epigenomic landscapes of pluripotent and lineage-committed human cells
- PMID: 20452322
- PMCID: PMC2867844
- DOI: 10.1016/j.stem.2010.03.018
Distinct epigenomic landscapes of pluripotent and lineage-committed human cells
Abstract
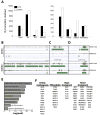
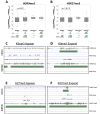
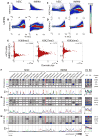
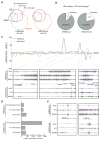
Human embryonic stem cells (hESCs) share an identical genome with lineage-committed cells, yet possess the remarkable properties of self-renewal and pluripotency. The diverse cellular properties in different cells have been attributed to their distinct epigenomes, but how much epigenomes differ remains unclear. Here, we report that epigenomic landscapes in hESCs and lineage-committed cells are drastically different. By comparing the chromatin-modification profiles and DNA methylomes in hESCs and primary fibroblasts, we find that nearly one-third of the genome differs in chromatin structure. Most changes arise from dramatic redistributions of repressive H3K9me3 and H3K27me3 marks, which form blocks that significantly expand in fibroblasts. A large number of potential regulatory sequences also exhibit a high degree of dynamics in chromatin modifications and DNA methylation. Additionally, we observe novel, context-dependent relationships between DNA methylation and chromatin modifications. Our results provide new insights into epigenetic mechanisms underlying properties of pluripotency and cell fate commitment.
Figures



References
-
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. - PubMed
-
- Barrand S, Collas P. Chromatin states of core pluripotency-associated genes in pluripotent, multipotent and differentiated cells. Biochem Biophys Res Commun. 2009;391:762–767. - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Ben-Shushan E, Pikarsky E, Klar A, Bergman Y. Extinction of Oct-3/4 gene expression in embryonal carcinoma x fibroblast somatic cell hybrids is accompanied by changes in the methylation status, chromatin structure, and transcriptional activity of the Oct-3/4 upstream region. Mol Cell Biol. 1993;13:891–901. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

