Genetic dissection of dietary restriction in mice supports the metabolic efficiency model of life extension
- PMID: 20452416
- PMCID: PMC2926251
- DOI: 10.1016/j.exger.2010.04.008
Genetic dissection of dietary restriction in mice supports the metabolic efficiency model of life extension
Abstract
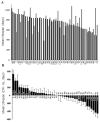
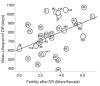
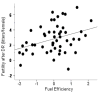
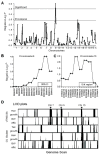
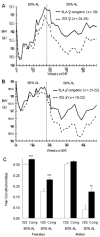
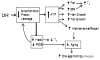
Dietary restriction (DR) has been used for decades to retard aging in rodents, but its mechanism of action remains an enigma. A principal roadblock has been that DR affects many different processes, making it difficult to distinguish cause and effect. To address this problem, we applied a quantitative genetics approach utilizing the ILSXISS series of mouse recombinant inbred strains. Across 42 strains, mean female lifespan ranged from 380 to 1070days on DR (fed 60% of ad libitum [AL]) and from 490 to 1020days on an AL diet. Longevity under DR and AL is under genetic control, showing 34% and 36% heritability, respectively. There was no correlation between lifespans on DR and AL; thus different genes modulate longevity under the two regimens. DR lifespans are significantly correlated with female fertility after return to an AL diet after various periods of DR (R=0.44, P=0.006). We assessed fuel efficiency (FE, ability to maintain growth and body weight independent of absolute food intake) using a multivariate approach and found it to be correlated with longevity and female fertility, suggesting possible causality. We found several quantitative trait loci responsible for these traits, mapping to chromosomes 7, 9, and 15. We present a metabolic model in which the anti-aging effects of DR are consistent with the ability to efficiently utilize dietary resources.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures

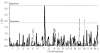
References
-
- Allison DB, Miller RA, Austad SN, Bouchard C, Leibel R, Klebanov S, Johnson TE, Harrison DE. Genetic variability in responses to caloric restriction in animals and in regulation of metabolism and obesity in humans. J Gerontol A Biol Sci Med Sci. 2001;56:55–65. - PubMed
-
- Belknap JK. Effect of within-strain sample size on QTL detection and mapping using recombinant inbred mouse strains. Behav Genet. 1998;28:29–38. - PubMed
-
- Belknap JK, Mitchell SR, O’Toole LA, Helms ML, Crabbe JC. Type I and type II error rates for quantitative trait loci (QTL) mapping studies using recombinant inbred mouse strains. Behav Genet. 1996;26:149–160. - PubMed
-
- Bennett B, Beeson M, Gordon L, Carosone-Link P, Johnson TE. Genetic dissection of quantitative trait loci specifying sedative/hypnotic sensitivity to ethanol: mapping with interval-specific congenic recombinant lines. Alcohol Clin Exp Res. 2002;26:1615–1624. - PubMed
-
- Chesler EJ, Miller DR, Branstetter LR, Galloway LD, Jackson BL, Philip VM, Voy BH, Culiat CT, Threadgill DW, Williams RW, Churchill GA, Johnson DK, Manly KF. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm Genome. 2008;19:382–389. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

