Human effector/initiator gene sets that regulate myometrial contractility during term and preterm labor
- PMID: 20452493
- PMCID: PMC2867841
- DOI: 10.1016/j.ajog.2010.02.034
Human effector/initiator gene sets that regulate myometrial contractility during term and preterm labor
Abstract
Objective: Distinct processes govern transition from quiescence to activation during term (TL) and preterm labor (PTL). We sought gene sets that are responsible for TL and PTL, along with the effector genes that are necessary for labor independent of gestation and underlying trigger.
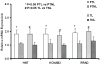
Study design: Expression was analyzed in term and preterm with or without labor (n=6 subjects/group). Gene sets were generated with logic operations.
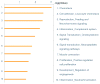
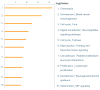
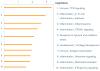
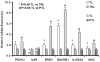
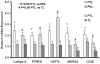
Results: Thirty-four genes were expressed similarly in PTL/TL but were absent from nonlabor samples (effector set); 49 genes were specific to PTL (preterm initiator set), and 174 genes were specific to TL (term initiator set). The gene ontogeny processes that comprise term initiator and effector sets were diverse, although inflammation was represented in 4 of the top 10; inflammation dominated the preterm initiator set.
Conclusion: TL and PTL differ dramatically in initiator profiles. Although inflammation is part of the term initiator and the effector sets, it is an overwhelming part of PTL that is associated with intraamniotic inflammation.
Copyright (c) 2010 Mosby, Inc. All rights reserved.
Figures




Similar articles
-
Immune cell and transcriptomic analysis of the human decidua in term and preterm parturition.Mol Hum Reprod. 2017 Oct 1;23(10):708-724. doi: 10.1093/molehr/gax038. Mol Hum Reprod. 2017. PMID: 28962035 Free PMC article.
-
Molecular signatures of labor and nonlabor myometrium with parsimonious classification from 2 calcium transporter genes.JCI Insight. 2021 Jun 8;6(11):e148425. doi: 10.1172/jci.insight.148425. JCI Insight. 2021. PMID: 33945511 Free PMC article.
-
Modeling hormonal and inflammatory contributions to preterm and term labor using uterine temporal transcriptomics.BMC Med. 2016 Jun 13;14(1):86. doi: 10.1186/s12916-016-0632-4. BMC Med. 2016. PMID: 27291689 Free PMC article.
-
Regulation of uterine contraction: mechanisms in preterm labor.AACN Clin Issues. 2000 May;11(2):271-82. doi: 10.1097/00044067-200005000-00010. AACN Clin Issues. 2000. PMID: 11235436 Review.
-
Mechanisms of full-term and preterm labor: factors influencing uterine activity.J Obstet Gynecol Neonatal Nurs. 1998 Nov-Dec;27(6):652-60. doi: 10.1111/j.1552-6909.1998.tb02635.x. J Obstet Gynecol Neonatal Nurs. 1998. PMID: 9836160 Review.
Cited by
-
Inflammatory gene regulatory networks in amnion cells following cytokine stimulation: translational systems approach to modeling human parturition.PLoS One. 2011;6(6):e20560. doi: 10.1371/journal.pone.0020560. Epub 2011 Jun 2. PLoS One. 2011. PMID: 21655103 Free PMC article.
-
Common mechanisms of physiological and pathological rupture events in biology: novel insights into mammalian ovulation and beyond.Biol Rev Camb Philos Soc. 2023 Oct;98(5):1648-1667. doi: 10.1111/brv.12970. Epub 2023 May 8. Biol Rev Camb Philos Soc. 2023. PMID: 37157877 Free PMC article. Review.
-
Transcription Analysis of the Myometrium of Labouring and Non-Labouring Women.PLoS One. 2016 May 13;11(5):e0155413. doi: 10.1371/journal.pone.0155413. eCollection 2016. PLoS One. 2016. PMID: 27176052 Free PMC article.
-
The role of the RHOA/ROCK pathway in the regulation of myometrial stages throughout pregnancy.AJOG Glob Rep. 2024 Sep 18;4(4):100394. doi: 10.1016/j.xagr.2024.100394. eCollection 2024 Nov. AJOG Glob Rep. 2024. PMID: 39434813 Free PMC article.
-
Evaluation of a Maternal Plasma RNA Panel Predicting Spontaneous Preterm Birth and Its Expansion to the Prediction of Preeclampsia.Diagnostics (Basel). 2022 May 27;12(6):1327. doi: 10.3390/diagnostics12061327. Diagnostics (Basel). 2022. PMID: 35741140 Free PMC article.
References
-
- Breuiller-Fouche M, Germain G. Gene and protein expression in the myometrium in pregnancy and labor. Reproduction. 2006;131:837–50. - PubMed
-
- Carbillon L, Seince N, Uzan M. Myometrial maturation and labour. Ann Med. 2001;33:571–8. - PubMed
-
- Norwitz ER, Robinson JN, Challis JR. The control of labor. N Engl J Med. 1999;341:660–6. - PubMed
-
- Buxton IL. Regulation of uterine function: a biochemical conundrum in the regulation of smooth muscle relaxation. Mol Pharmacol. 2004;65:1051–9. - PubMed
-
- Hillhouse EW, Grammatopoulos DK. Role of stress peptides during human pregnancy and labour. Reproduction. 2002;124:323–9. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

