Protein subcellular localization in bacteria
- PMID: 20452938
- PMCID: PMC2845201
- DOI: 10.1101/cshperspect.a000307
Protein subcellular localization in bacteria
Abstract
Like their eukaryotic counterparts, bacterial cells have a highly organized internal architecture. Here, we address the question of how proteins localize to particular sites in the cell and how they do so in a dynamic manner. We consider the underlying mechanisms that govern the positioning of proteins and protein complexes in the examples of the divisome, polar assemblies, cytoplasmic clusters, cytoskeletal elements, and organelles. We argue that geometric cues, self-assembly, and restricted sites of assembly are all exploited by the cell to specifically localize particular proteins that we refer to as anchor proteins. These anchor proteins in turn govern the localization of a whole host of additional proteins. Looking ahead, we speculate on the existence of additional mechanisms that contribute to the organization of bacterial cells, such as the nucleoid, membrane microdomains enriched in specific lipids, and RNAs with positional information.
Figures

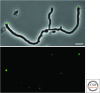

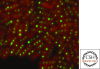
References
-
- Alley MR, Maddock JR, Shapiro L 1992. Polar localization of a bacterial chemoreceptor. Genes Dev 6:825–836 - PubMed
-
- Arigoni F, Pogliano K, Webb CD, Stragier P, Losick R 1995. Localization of protein implicated in establishment of cell type to sites of asymmetric division. Science 270:637–640 - PubMed
-
- Ausmees N, Kuhn JR, Jacobs-Wagner C 2003. The bacterial cytoskeleton: An intermediate filament-like function in cell shape. Cell 115:705–713 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
