ZAP-70: an essential kinase in T-cell signaling
- PMID: 20452964
- PMCID: PMC2857167
- DOI: 10.1101/cshperspect.a002279
ZAP-70: an essential kinase in T-cell signaling
Abstract
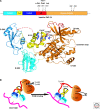
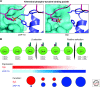
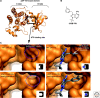
ZAP-70 is a cytoplasmic protein tyrosine kinase that plays a critical role in the events involved in initiating T-cell responses by the antigen receptor. Here we review the structure of ZAP-70, its regulation, its role in development and in disease. We also describe a model experimental system in which ZAP-70 function can be interrupted by a small chemical inhibitor.
Figures
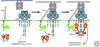

References
-
- Allsup DJ, Kamiguti AS, Lin K, Sherrington PD, Matrai Z, Slupsky JR, Cawley JC, Zuzel M 2005. B-cell receptor translocation to lipid rafts and associated signaling differ between prognostically important subgroups of chronic lymphocytic leukemia. Cancer Res 65:7328–7337 - PubMed
-
- Arpaia E, Shahar M, Dadi H, Cohen A, Roifman CM 1994. Defective T cell receptor signaling and CD8+ thymic selection in humans lacking zap-70 kinase. Cell 76:947–958 - PubMed
-
- Au-Yeung BB, Deindl S, Hsu LY, Palacios EH, Levin SE, Kuriyan J, Weiss A 2009. The structure, regulation, and function of ZAP-70. Immunol Rev 228:41–57 - PubMed
-
- Bishop AC, Shah K, Liu Y, Witucki L, Kung C, Shokat KM 1998. Design of allele-specific inhibitors to probe protein kinase signaling. Curr Biol 8:257–266 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
