p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription
- PMID: 20452972
- PMCID: PMC2903417
- DOI: 10.1074/jbc.M110.118976
p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription
Abstract
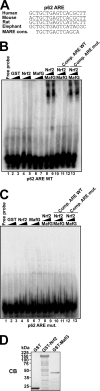
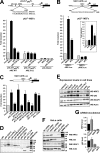
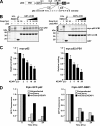
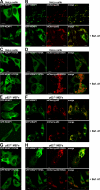
The p62/SQSTM1 (sequestosome 1) protein, which acts as a cargo receptor for autophagic degradation of ubiquitinated targets, is up-regulated by various stressors. Induction of the p62 gene by oxidative stress is mediated by NF-E2-related factor 2 (NRF2) and, at the same time, p62 protein contributes to the activation of NRF2, but hitherto the mechanisms involved were not known. Herein, we have mapped an antioxidant response element (ARE) in the p62 promoter that is responsible for its induction by oxidative stress via NRF2. Chromatin immunoprecipitation and gel mobility-shift assays verified that NRF2 binds to this cis-element in vivo and in vitro. Also, p62 docks directly onto the Kelch-repeat domain of Kelch-like ECH-associated protein 1 (KEAP1), via a motif designated the KEAP1 interacting region (KIR), thereby blocking binding between KEAP1 and NRF2 that leads to ubiquitylation and degradation of the transcription factor. The KIR motif in p62 is located immediately C-terminal to the LC3-interacting region (LIR) and resembles the ETGE motif utilized by NRF2 for its interaction with KEAP1. KIR is required for p62 to stabilize NRF2, and inhibition of KEAP1 by p62 occurs from a cytoplasmic location within the cell. The LIR and KIR motifs cannot be engaged simultaneously by LC3 and KEAP1, but because p62 is polymeric the interaction between KEAP1 and p62 leads to accumulation of KEAP1 in p62 bodies, which is followed by autophagic degradation of KEAP1. Our data explain how p62 contributes to activation of NRF2 target genes in response to oxidative stress through creating a positive feedback loop.
Figures


References
-
- Halliwell B., Gutteridge J. M. (eds) (2007) Free Radicals in Biology and Medicine, 4th Ed., pp. 187–267, Oxford University Press, Oxford, UK
-
- Hayes J. D., McMahon M. (2009) Trends Biochem. Sci. 34, 176–188 - PubMed
-
- Kensler T. W., Wakabayashi N., Biswal S. (2007) Annu. Rev. Pharmacol. Toxicol. 47, 89–116 - PubMed
-
- Motohashi H., Yamamoto M. (2004) Trends Mol. Med. 10, 549–557 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

