Recombination-associated sequence homogenization of neighboring Alu elements: signature of nonallelic gene conversion
- PMID: 20453015
- PMCID: PMC2950799
- DOI: 10.1093/molbev/msq116
Recombination-associated sequence homogenization of neighboring Alu elements: signature of nonallelic gene conversion
Abstract
Recently, researchers have begun to recognize that, in order to establish neutral models for disease association and evolutionary genomics studies, it is crucial to have a clear understanding of the genomic impact of nonallelic gene conversion. Drawing on previous successes in characterizing this phenomenon over protein-coding gene families, we undertook a computational analysis of neighboring Alu sequences in the genome scale. For this purpose, we developed adjusted comutation rate (aCMR), a novel statistical method measuring the excess number of identical point mutations shared by adjacent Alu sequences, vis-à-vis random pairs. Using aCMR, we uncovered a remarkable genome-wide sequence homogenization of neighboring Alus, with the strongest signal observed in the pseudoautosomal regions of the X and Y chromosomes. The magnitude of sequence homogenization between Alu pairs is greater with shorter interlocus distance, higher sequence identity, and parallel orientation. Moreover, shared substitutions show a strong directionality toward GC nucleotides, with multiple substitutions tending to cluster within the Alu sequence. Taken together, these observed recombination-associated sequence homogenization patterns are best explained by frequent ubiquitous gene conversion events between neighboring Alus. We believe that these observations help to illuminate the nature and impact of the enigmatic phenomenon of gene conversion.
Figures
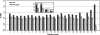


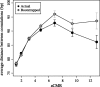
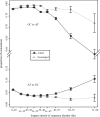
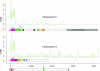
Similar articles
-
Biased distribution of inverted and direct Alus in the human genome: implications for insertion, exclusion, and genome stability.Genome Res. 2001 Jan;11(1):12-27. doi: 10.1101/gr.158801. Genome Res. 2001. PMID: 11156612
-
Potential gene conversion and source genes for recently integrated Alu elements.Genome Res. 2000 Oct;10(10):1485-95. doi: 10.1101/gr.152300. Genome Res. 2000. PMID: 11042148
-
Non-traditional Alu evolution and primate genomic diversity.J Mol Biol. 2002 Mar 8;316(5):1033-40. doi: 10.1006/jmbi.2001.5380. J Mol Biol. 2002. PMID: 11884141
-
Alu repeats and human genomic diversity.Nat Rev Genet. 2002 May;3(5):370-9. doi: 10.1038/nrg798. Nat Rev Genet. 2002. PMID: 11988762 Review.
-
Alu elements and the human genome.Genetica. 2000;108(1):57-72. doi: 10.1023/a:1004099605261. Genetica. 2000. PMID: 11145422 Review.
Cited by
-
Gene Conversion amongst Alu SINE Elements.Genes (Basel). 2021 Jun 11;12(6):905. doi: 10.3390/genes12060905. Genes (Basel). 2021. PMID: 34208107 Free PMC article.
-
Structural variation and its potential impact on genome instability: Novel discoveries in the EGFR landscape by long-read sequencing.PLoS One. 2020 Jan 15;15(1):e0226340. doi: 10.1371/journal.pone.0226340. eCollection 2020. PLoS One. 2020. PMID: 31940362 Free PMC article.
-
Population-specific differences in gene conversion patterns between human SUZ12 and SUZ12P are indicative of the dynamic nature of interparalog gene conversion.Hum Genet. 2014 Apr;133(4):383-401. doi: 10.1007/s00439-013-1410-4. Epub 2014 Jan 3. Hum Genet. 2014. PMID: 24385046
-
The Role of Gene Conversion between Transposable Elements in Rewiring Regulatory Networks.Genome Biol Evol. 2019 Jul 1;11(7):1723-1729. doi: 10.1093/gbe/evz124. Genome Biol Evol. 2019. PMID: 31209488 Free PMC article. Review.
-
Significant selective constraint at 4-fold degenerate sites in the avian genome and its consequence for detection of positive selection.Genome Biol Evol. 2011;3:1381-9. doi: 10.1093/gbe/evr112. Epub 2011 Oct 31. Genome Biol Evol. 2011. PMID: 22042333 Free PMC article.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Arndt PF, Petrov DA, Hwa T. Distinct changes of genomic biases in nucleotide substitution at the time of Mammalian radiation. Mol Biol Evol. 2003;20:1887–1896. - PubMed
-
- Benovoy D, Drouin G. Ectopic gene conversions in the human genome. Genomics. 2009;93:27–32. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

