Can a sex-biased human demography account for the reduced effective population size of chromosome X in non-Africans?
- PMID: 20453016
- PMCID: PMC2944028
- DOI: 10.1093/molbev/msq117
Can a sex-biased human demography account for the reduced effective population size of chromosome X in non-Africans?
Abstract
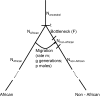
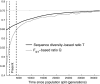
Sex-biased demographic events can result in asymmetries in female and male effective population size that can lead to different patterns of genetic variation on chromosome X than are expected based on the patterns on the autosomes. Previous studies point to a period around the time of the dispersal of anatomically modern humans out of Africa when chromosome X experienced a significant reduction in effective population size relative to the autosomes. Here, we explore whether a sex-biased demographic history could explain these observations. We use coalescent simulations to show that a model of primarily male migration during the out-of-Africa dispersal can produce the striking patterns that are observed when comparing patterns of genetic variation on the autosomes and chromosome X. The model involves a history in which after the founder population of non-Africans lost much of its genetic diversity, subsequent mostly male gene flow from an African source brought new diversity into the population. We also explore two additional models, one of sex-biased generation time and one of a substructured population during the dispersal out of Africa with primarily female migration among demes. These latter models cannot account for the magnitude of the observed reduction in chromosome X effective population size, although it is plausible that they played a more minor role in producing the striking chromosome X/autosome patterns.
Figures
Similar articles
-
Accelerated genetic drift on chromosome X during the human dispersal out of Africa.Nat Genet. 2009 Jan;41(1):66-70. doi: 10.1038/ng.303. Epub 2008 Dec 21. Nat Genet. 2009. PMID: 19098910 Free PMC article.
-
The inference of sex-biased human demography from whole-genome data.PLoS Genet. 2019 Sep 20;15(9):e1008293. doi: 10.1371/journal.pgen.1008293. eCollection 2019 Sep. PLoS Genet. 2019. PMID: 31539367 Free PMC article.
-
Sex-biased evolutionary forces shape genomic patterns of human diversity.PLoS Genet. 2008 Sep 26;4(9):e1000202. doi: 10.1371/journal.pgen.1000202. PLoS Genet. 2008. PMID: 18818765 Free PMC article.
-
Looking for signatures of sex-specific demography and local adaptation on the X chromosome.Genome Biol. 2010 Jan 28;11(1):203. doi: 10.1186/gb-2010-11-1-203. Genome Biol. 2010. PMID: 20236443 Free PMC article. Review.
-
The different levels of genetic diversity in sex chromosomes and autosomes.Trends Genet. 2009 Jun;25(6):278-84. doi: 10.1016/j.tig.2009.04.005. Epub 2009 May 27. Trends Genet. 2009. PMID: 19481288 Review.
Cited by
-
Strong Selective Sweeps on the X Chromosome in the Human-Chimpanzee Ancestor Explain Its Low Divergence.PLoS Genet. 2015 Aug 14;11(8):e1005451. doi: 10.1371/journal.pgen.1005451. eCollection 2015 Aug. PLoS Genet. 2015. PMID: 26274919 Free PMC article.
-
Identifying X-chromosome variants associated with age-related macular degeneration.Hum Mol Genet. 2024 Dec 6;33(24):2085-2093. doi: 10.1093/hmg/ddae141. Hum Mol Genet. 2024. PMID: 39324238 Free PMC article.
-
The Nubian Complex of Dhofar, Oman: an African middle stone age industry in Southern Arabia.PLoS One. 2011;6(11):e28239. doi: 10.1371/journal.pone.0028239. Epub 2011 Nov 30. PLoS One. 2011. PMID: 22140561 Free PMC article.
-
Extensive X-linked adaptive evolution in central chimpanzees.Proc Natl Acad Sci U S A. 2012 Feb 7;109(6):2054-9. doi: 10.1073/pnas.1106877109. Epub 2012 Jan 23. Proc Natl Acad Sci U S A. 2012. PMID: 22308321 Free PMC article.
-
Changes in life history and population size can explain the relative neutral diversity levels on X and autosomes in extant human populations.Proc Natl Acad Sci U S A. 2020 Aug 18;117(33):20063-20069. doi: 10.1073/pnas.1915664117. Epub 2020 Aug 3. Proc Natl Acad Sci U S A. 2020. PMID: 32747577 Free PMC article.
References
-
- Bar-Yosef O, Belfer-Cohen A. The Levantine “PPNB” interaction sphere. In: Hershkovitz I, editor. People and culture in change. Oxford: British Archaeological Reports; 1989. pp. 59–72.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

